INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China in December 2019, causing asymptomatic or mild to severe disease of respiratory tract, coronavirus disease 2019 (COVID-19) [1]. As of August 6, 2020, the pandemic of COVID-19 has spread to 133 countries with >18 million-confirmed cases and >700,000 confirmed deaths [2].
SARS-CoV-2 real-time reverse transcription polymerase chain reaction (rRT-PCR) is the standard test to diagnose COVID-19, as recommended by World Health Organization (WHO) [3]. As of July 16, 2020, the Foundation for Innovative New Diagnostics (FIND) listed 347 commercial molecular assays available worldwide (www.finddx.org/covid-19/pipeline on July 16, 2020). Until July 26, 2020, 16 molecular diagnostic assays for SARS-CoV-2 have been approved under Emergency Use Authorizations (EUAs) to respond to COVID-19 pandemic in Korea [4].
SARS-CoV-2 rRT-PCR is recommended to design using multiple target genes with the open reading frame 1ab/ RNA-dependent RNA polymerase (ORF1ab/RdRp), envelope (E), nucleocapsid (N), and spike (S) genes [5]. According to the guidance of the Korea Centers for Disease Control (KCDC) as well as the WHO, all target genes in PCR kits should be positive for laboratory diagnosis of COVID-19 [3,6]. However, SARS-CoV-2 rRT-PCR can provide inconclusive results when, one or more, but not all of multiple target genes are positive, thereby creating difficulties for COVID-19 diagnosis in clinical laboratories [7,8].
Inconclusive results could be caused by various factors from contamination to mutation of SARS-CoV-2 [7,9]. Moreover, the clinical relevance of inconclusive results is not well understood. It is important how to interpret such results, since the SARS-CoV-2 rRT-PCR data is implied as indicator of the infectivity and the necessity of quarantine in COVID-19 patients. The purpose of this study is to investigate the causes and clinical relevance of inconclusive results of SARS-CoV-2 rRT-PCR.
MATERIALS AND METHOD
For a four-month period from March to June, 2020, SARS-CoV-2 rRT-PCR results were retrospectively reviewed in the clinical microbiology laboratory based on a tertiary care hospital of 2,705 beds in Seoul, Korea. SARS-CoV-2 rRT-PCR was performed using the Allplex 2019-nCoV assay kit (Seegene Inc., Seoul, Korea) to detect three target genes, E, RdRp, and N. This assay kit received a Conformité Européenne (CE) marking as an in vitro Diagnostic (IVD) device on February 10, 2020 and Emergency Use Authorization (EUA) by the Ministry of Food and Drug Safety on February 12, 2020.
The SARS-CoV-2 rRT-PCR with Allplex is interpret positive when the cycle threshold (Ct) values of all target genes are within 40. Therefore, inconclusive interpretations are made when only 1-2 target genes are amplified with the Ct values of less than 40 [6].
All inconclusive test results were analyzed for specimen types, the Ct and peak values of amplification curves, the positive target genes, and the results of repeat or consecutive tests for the patient with inconclusive tests. Frequency of inconclusive tests and Ct values of E gene of positive control were analyzed by each kit lot number. For statistical analysis, t-tests or Mann-Whitney U test were applied to comparison of the Ct values and relative fluorescence units (RFUs) of peaks of positive target genes between COVID-19 and non-COVID-19 cases, and frequency of inconclusive results per the kit lot number. The average Ct values of E gene of positive control were compared using Kruskal-Wallis test between each lot number of kits. A P value < 0.05 was regarded as the threshold for statistical significance. Statistical analyses were conducted using MedCalc (version 19.2.1; MedCalc Software, Ostend, Belgium).
This study was approved by the Institutional Review Board of Asan Medical Center, Seoul, Korea and was deemed to be exempt from informed consent (IRB No. 2020-1444).
RESULTS
During the study period, a total of 43,268 SARS-CoV-2 rRT-PCR tests including 39,998 (92.4%) nasopharyngeal swab samples and 3,270 (7.6%) sputum samples were conducted. At that time, 14 cases of COVID-19 were diagnosed and 93 (0.21%) inconclusive results were detected in the laboratory. Of the inconclusive results, 49 (52.7%) corresponded to the tests of 11 laboratory-confirmed COVID-19 patients, and 44 (47.3%) were of non-COVID-19 patients (Fig. 1). The specimens of inconclusive results comprised 31 nasopharyngeal swab samples and 18 sputum samples in COVID-19 patients, and 38 nasopharyngeal swab samples and 6 sputum samples in non-COVID-19 patients. The inconclusive results from COVID-19 patients were usually detected 11.9 ± 4.7 days after the diagnosis of COVID-19, except in cases of initially inconclusive results such as in one past COVID-19 patient (patient 8) and one newly diagnosed COVID-19 patient (patient 11) (Table 1). The patient 8 was diagnosed COVID-19 68 days ago and converted negative one month before admission in our hospital, who showed inconclusive results twice on the 8th and 26th hospital days. The patient 11 was diagnosed at the COVID-19 screening center of our hospital, which was confirmed positive by resampling after initial inconclusive result. From the time of the initial inconclusive result, it took 8.8 ± 5.5 days to satisfy the criterion of negative conversion, i.e. two consecutive negative PCR test results obtained at least 24 hours apart [10] (Table 1). During this period, there were 3.5 ± 3.0 positive, 3.8 ± 2.4 negative, and 3.0 ± 2.1 inconclusive results in consecutive tests. Two patients (patient 1 and patient 3) that did not receive any antiviral treatment showed a self-limiting course of COVID-19, with the SARS-CoV-2 rRT-PCR tests producing gradually increasing Ct values followed by inconclusive results, and eventually converting to negative 1 month after diagnosis. Fig. 2A showed the trend of the consecutive tests of the patient 3. The longest spanning period of inconclusive results occurred in a severe case of a COVID-19 (patient 4) who was treated with extracorporeal membrane oxygenation and low doses of steroids [11] and her nasopharyngeal swab samples produced 5 inconclusive results in a period of 30 days (Fig. 2B). Two other patients were not followed up until negative conversion because one patient was transferred to another hospital and the other patient, diagnosed at screening center, was admitted to a COVID-19-dedicated hospital. Furthermore, among 3 COVID-19 patients not displaying inconclusive results, two were transferred to other hospitals when SARS-CoV-2 rRT-PCR tests were still showing positive results, and one was diagnosed only on June 29 (Table 1).
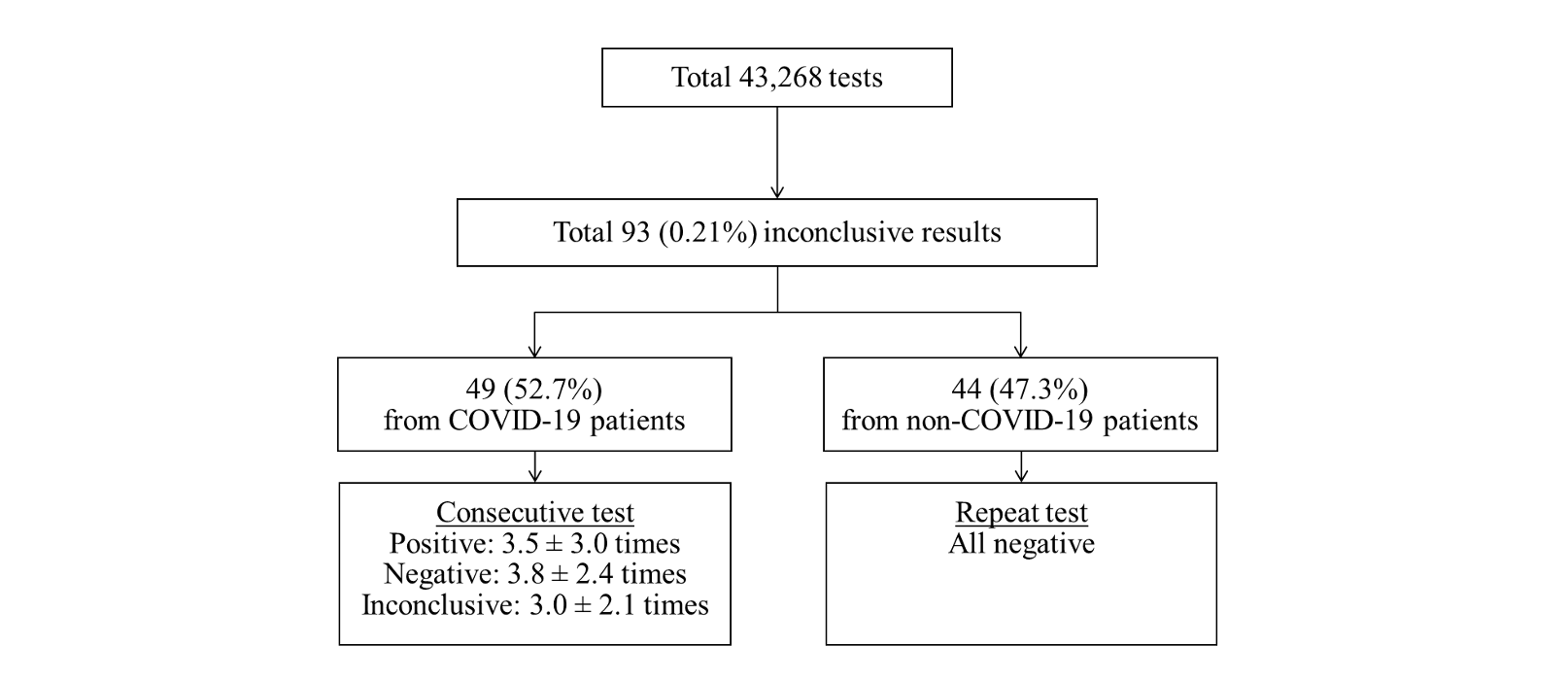
Fig. 1. Classification of the inconclusive results according to COVID-19 diagnosis and retest or consecutive test results.
Abbreviation: COVID-19, coronavirus disease 19.
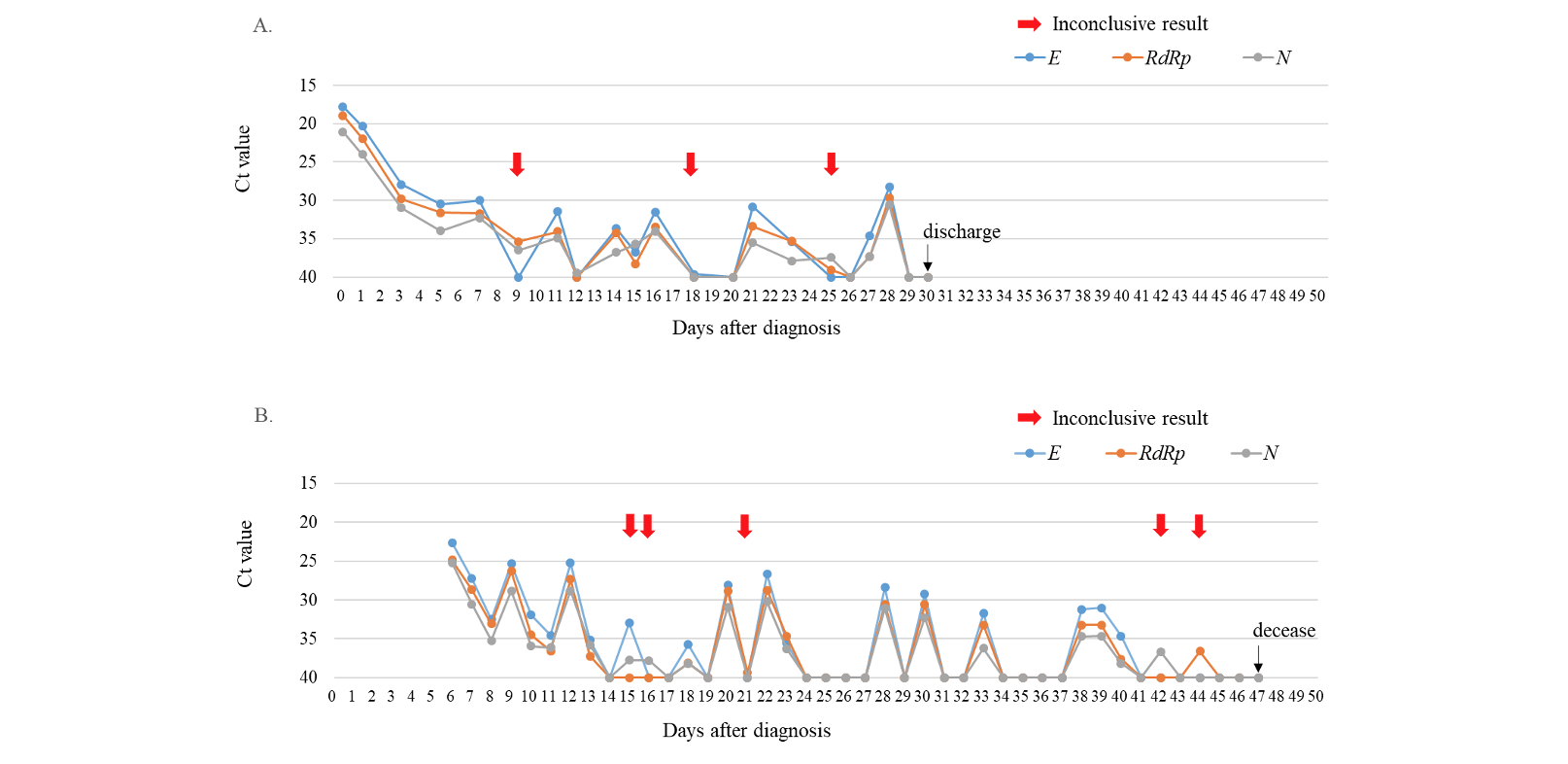
Fig. 2. Trends of Ct values of three target genes when consecutive SARS-CoV-2 rRT-PCR tests were conducted with nasopharyngeal swab samples from two COVID-19 patients. (A) The asymptomatic patient without treatment (patient 3). (B) The patient with severe symptoms under extracorporeal membrane oxygenation and low-dose steroid treatments (patient 4). Red arrows indicated inconclusive results.
Abbreviations: Ct, cycle threshold; E, envelope gene; RdRp, RNA-dependent RNA polymerase gene; N, nucleocapsid gene.
Table 1
Summary of rRT-PCR test producing inconclusive results from 11 COVID-19 patients
*Drop out because transferred to another hospital before negative conversion.
†Diagnosis was made in retesting on same day and there was no further test owing to admission to another hospital.
Abbreviations: rRT-PCR, real time reverse transcription polymerase chain reaction; COVID-19, coronavirus disease 2019; Ct, threshold cycle; RdRp, RNA-dependent RNA polymerase gene; E, envelope gene; N, nucleocapsid gene; N/A, not applicable/available; ND, not detected; Pos, positive test; Inc, inconclusive test; Neg, negative test; ECMO, extracorporeal membrane oxygenation; HCQ, hydroxychloroquine
Notably, all inconclusive specimens from non-COVID-19 patients were negative upon retest. Of 8 lot of Allplex kits used during study period, two lot numbers, RP4520C20 and RP4520C27, yielded 36 (81.8%) inconclusive results, which was significantly more frequent than the other 6 lots (Table 2). The average Ct values of E gene of positive control were from 19.02 ± 0.41 to 19.98 ± 0.45. The lot of RP4520C20 showed the average of 19.97 ± 0.27, similar to 19.97 ± 0.20 in RP4520D68 and longer than those of RP4520C20 (P < 0.01). Therefore, a difference in Ct value of positive control was not relative to number of inconclusive results per lot. On the other hand, the inconclusive results occurred in COVID-19 patients were not relative to specific lot number. The average Ct value of positive target genes was 37.3 ± 2.0 and all Ct values were ≥ 33 except 4 samples from non-COVID-19 cases (Table 3).
Table 2
Lot number of tested kits and results of retesting of samples in non-COVID-19 patients displaying inconclusive results
Table 3
Comparison of frequency and Ct and peak value of amplification curves of positive target genes in inconclusive results between COVID-19 cases and non-COVID-19 cases
The most frequent positive target gene were N (65.3%) in COVID-19 patients’ samples, and RdRp (61.2%) in non-COVID-19 patients’ samples, respectively. Positive results for two target genes were found in 36.7% of inconclusive results of COVID-19 patients and not in non-COVID-19 patients. However, Ct values and peak RFUs were not significantly different between the two groups (P > 0.05, respectively) (Table 3).
DISCUSSION
Inconclusive results when testing samples from COVID-19 patients, were produced 11.9 ± 4.7 days after diagnosis of COVID-19. The Ct values of target genes relative to these inconclusive results were all above 33. Previous studies have shown that the Ct value of positive target genes in SARS CoV-2 rRT-PCR is well correlated with the viral load [8,12]. Therefore, inconclusive results from COVID-19 patients may indicate that the viral load decreased during recovery from COVID-19. Thus, inconclusive results could derive from a natural process during follow-up of patients with positive SARS-CoV-2 rRT-PCR. Indeed, as shown by “re-positive” cases, the RNA of SARS-CoV-2 can be detected long after the disappearance of virus infectivity [13,14]; therefore, inconclusive results associated with a lower viral load than positive results may indicate no infectivity. A symptom-based and a time-based strategy to release quarantine is added to the previous test-based strategy on June 27, 2020 in Korea [10,15,16]. With a time-based strategy, quarantine can be released 10 days after first COVID-19 positive test if there is no subsequent illness whether or not positive in rRT-PCR, which is based on the assumption of no more infectivity in that condition. The finding that on average 12 days after diagnosis yielded inconclusive results was consistent with this time-based strategy. However, inconclusive results were often followed by positive results in this study. Thus, inconclusive results should not be counted as positive or negative for applying test-based strategy.
All inconclusive results in non-COVID-19 cases were single gene-positive and were negative upon retest, thus false positive, and inconclusive results was rather associated with the specific lot of the kits and positive RdRp. Indeed, unequal distribution of inconclusive results between the lots could not be explained only by contamination. This finding suggested that kits of certain lot number could be predisposed to produce false positive for certain target genes. However, lack of correlation between Ct value of the positive control and frequency of inconclusive tests per lot suggested that the lot to lot variation of PCR efficiency did not explain occurrence of false positives. This kind of lot problem could be solved by changing the lot [7]. Because false positive is the most frequent cause of inconclusive result in this study, retesting is necessary when inconclusive results are initially encountered in non-COVID-19 or newly diagnosed patients. There was a serious concern that inconclusive results could be due to mutations of target genes of SARS-CoV-2, which would be a caveat for the performance of primers or probes in rRT-PCR [9]. However, in this study, the COVID-19 cases repeatedly showing inconclusive results had consecutive samples which were positive for alternating target genes upon retest. Therefore, mutation was unlikely to have caused inconclusive results in this study.
Among target genes of SARS-COV-2, the relative abundance of N gene subgenomic mRNA produced during virus replication [17] could theoretically enhance diagnostic sensitivity of N gene, and other study showed that the N gene-based rRT-PCR assay was more sensitivity than other target gene assay for detection of SARS-CoV-2 [18,19]. This may explain why N was the most frequent target gene to be positive in the inconclusive results encountered in COVID-19 patients. In contrast, in non-COVID-19 patients, all inconclusive results came from single target gene-positive and RdRp was the most frequently positive as an only one positive target gene. Although association of specific lot to false positive RdRp has not been reported elsewhere [7], occurrence of inconclusive results could depend on quality control issue in lots or shipments of the specific brand kits. Therefore, close monitoring of lot changes or shipments would be incorporated to the quality control plan of SARS-CoV-2 rRT-PCR.
There was no difference of Ct value or peak RFU between the inconclusive results from COVID-19 and non-COVID-19 cases. Thus, the analysis of amplification curves of the positive target genes was not helpful to distinguish inconclusive results due to low viral load in COVID-19 from false positive in non-COVID-19 patients. Therefore, inconclusive results in non-COVID-19 patients or newly diagnosed patient required confirmatory retesting. Nevertheless double gene positivity raises suspicion that it may be COVID-19, because all false positives are obtained with a single gene. Thus, testing three target genes in a multiplex PCR system such as Allplex could be advantageous.
Our study has some limitations. First, this study was carried out with a small number of COVID-19 patients. Therefore, further study is required with more COVID-19 cases to assess the risk of various potential cause of inconclusive results, such as mutation of target genes. Secondly, only specific lot or shipment of assay kits was noted as a source of the inconclusive results in other than COVID-19. To complete quality control plan, the risk of laboratory contamination or technical failure to detect all positive target genes should be assessed in own SARS-COV-2 rRT-PCR system of each laboratory.
In conclusion, inconclusive results could derive from the natural process of deceasing in viral load in COVID-19 patients, while they mostly represent false positives in non-COVID-19 patients. Therefore, we suggested the guidance for reporting inconclusive results that it could be reported as inconclusive without retesting in confirmed COVID-19 patients, but should be retested for non-COVID-19 patients or newly diagnosed patients.




