Abstract
Background: The prevalence of carbapenemase-producing Enterobacteriaceae (CPE), especially the KPC-2-producing Klebisella pneumoniae, is rapidly increasing and becoming a menace to global public health. This study aims to present the molecular epidemiology of the KPC-2-producing K. pneumoniae isolates emerged in a tertiary hospital in South Korea and describe its clinical significance. Methods: This study included carbapenem-resistant K. pneumoniae isolates collected from a tertiary hospital from April to December in 2018. Antimicrobial susceptibility of K. pneumoniae isolates was tested using disk diffusion method. PCR and DNA sequence analyses were performed to identify the resistance genotype. In addition, the molecular epidemiology was investigated using pulsed-field gel electrophoresis (PFGE) and multilocus sequencing typing (MLST). Results: Total 100 KPC-2-producing K. pneumoniae isolates were collected, which were mainly classified into two pulsotypes according to the XbaI restriction digestion pattern by PGFE analysis (pulsotype A, n = 31; pulsotype B, n = 63). The isolates exhibiting pulsotype A belonged to ST395 and the remaining isolates exhibiting pulsotype B were attributed to ST307 by MLST analysis. Conclusion: This study investigated clinical information and molecular bacterial profiles for KPC-2-producing K. pneumoniae isolates. These findings indicate that the proper infection control activities are needed to prevent the spread of multidrug-resistant organisms such as CPE, which could cause high mortality in clinical field.
Keywords
Carbapenemase KPC Klebsiella pneumoniae PFGE
Figures & Tables
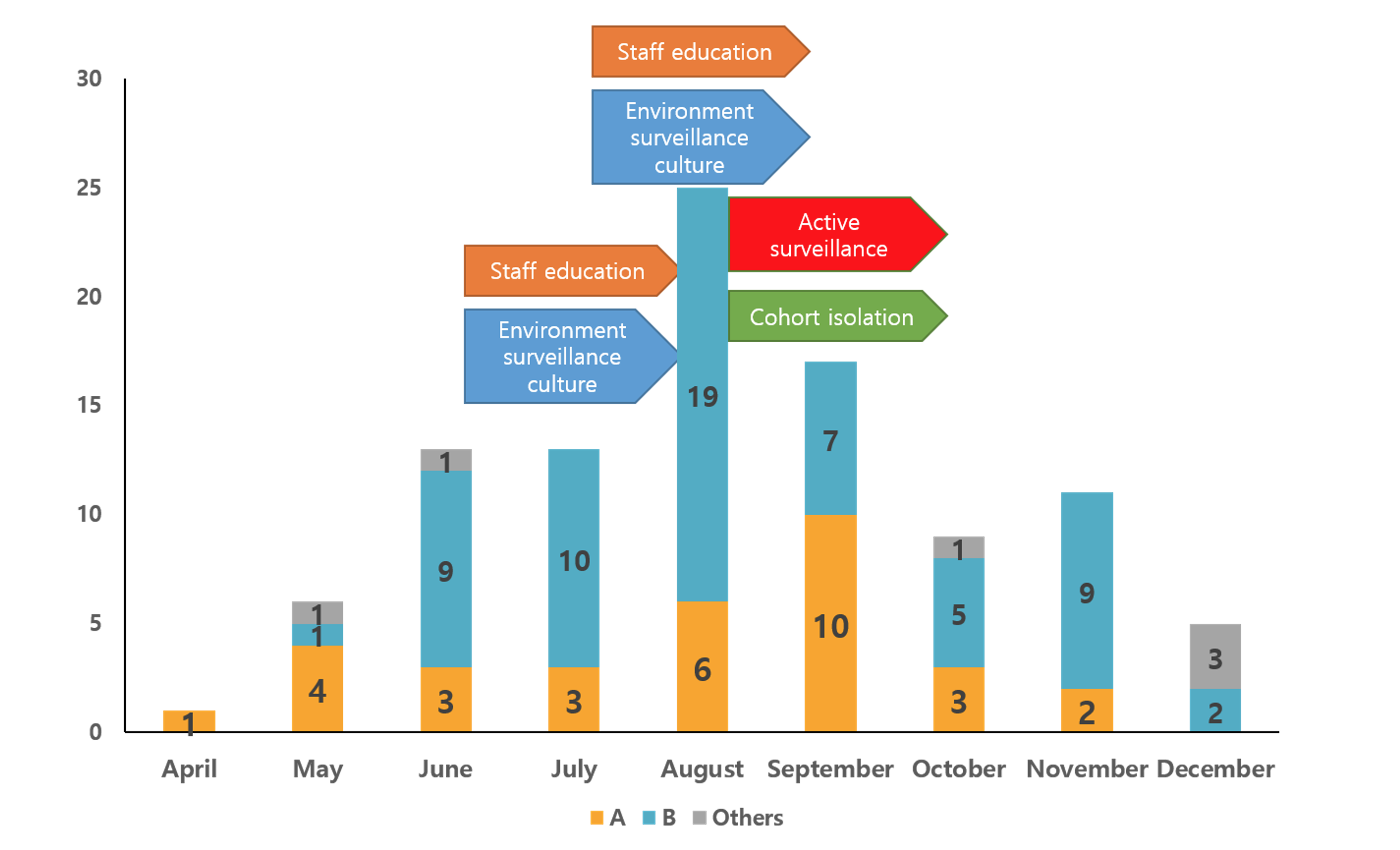
Figure 1. Monthly isolation of KPC-2 producing K. pneumoniae isolates stratified by PFGE banding patterns. Bar graphs indicate the numbers of KPC-2 producing K. pneumoniae isolates collected in each nine month in 2018 and arrows indicate the activity performed for eradicating a KPC-type carbapenemase producing K. pneumoniae isolates outbreak. Yellow bar, pulsotype A; Blue bar, pulsotype B; Gray bar, other pulsotypes.
Figures & Tables
Table 1. Oligonucleotide sequence of the primers used in this study
| Target gene | Primer name | Primer sequence (5' to 3') | Amplicon size (bp) |
| blaCTX-M-1 | CTX-M-1_-48–25F | GACTATTCATGTTGTTGTTAWTTC | 973 |
| CTX-M-1_+28-+49R | TAAGGCGATAAACAAAAACGGA | ||
| blaCTX-M-9 | CTX-M-9_-42—21F | GAATACTGATGTAACACGGATT | 962 |
| CTX-M-9-1_+23-+44R | ATATAAATAGAAAGTGGGGCAC | ||
| CTX-M-9-2_+27-+46R | CTGATCCTTCAACTCAGCAA | ||
| blaCMY | MOXM F | GCTGCTCAAGGAGCACAGGAT | 520 |
| MOXM R | CACATTGACATAGGTGTGGTGC | ||
| blaCMY-2 | CITM F | TGGCCAGAACTGACAGGCAAA | 462 |
| CITM R | TTTCTCCTGAACGTGGCTGGC | ||
| blaDHA | DHAM F | AACTTTCACAGGTGTGCTGGGT | 405 |
| DHAM R | CCGTACGCATACTGGCTTTGC | ||
| blaACC | ACCM F | AACAGCCTCAGCAGCCGGTTA | 346 |
| ACCM R | TTCGCCGCAATCATCCCTAGC | ||
| blaACT | EBCM F | TCGGTAAAGCCGATGTTGCGG | 302 |
| EBCM R | CTTCCACTGCGGCTGCCAGTT | ||
| blaFOX | FOXM F | AACATGGGGTATCAGGGAGATG | 190 |
| FOXM R | CAAAGCGCGTAACCGGATTGG | ||
| blaKPC | KPC F | ATGTCACTGTATCGCCGTCT | 882 |
| KPC R | TTTTCAGAGCCTTACTGCCC |


