Department of Laboratory Medicine, Chonnam National University Medical School, Gwangju, Korea
Corresponding to Jong Hee Shin, E-mail: shinjh@chonnam.ac.kr
Ann Clin Microbiol 2024;27(4):231-244. https://doi.org/10.5145/ACM.2024.27.4.3
Received on 9 October 2024, Revised on 27 November 2024, Accepted on 29 November 2024, Published on 20 December 2024.
Copyright © Korean Society of Clinical Microbiology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND) (https://creativecommons.org/licenses/by-nc-nd/4.0/).
Purpose: Candidemia is a common cause of nosocomial bloodstream infections associated with high mortality rates. Its incidence varies significantly across countries and hospitals, and its epidemiology is a subject of continuous investigation. This review aims to provide a comprehensive analysis of candidemia in Korea, addressing its changing epidemiology, species distribution, antifungal resistance, and clinical implications. Current content: In Korea, Candida albicans remains the most common isolate in blood cultures; however, infections caused by non-albicans Candida species are increasing. The 30day mortality rates for patients with candidemia vary considerably across different Candida species, with Candida tropicalis at 47.0%, C. albicans at 36.4%, Candida glabrata at 34.7%, and Candida parapsilosis at 22.5%. Recent Korean studies have highlighted the clonal spread of bloodstream infections caused by C. parapsilosis with the Erg11p Y132F mutation, and certain isolates are becoming endemic to specific healthcare settings. C. glabrata poses a significant threat; this species is increasingly resistant to antifungal medications and multidrug-resistant isolates are emerging. Whole-genome sequencing analysis elucidates the transmission dynamics of clonal bloodstream isolates of C. glabrata among patients receiving antifungal therapy. This analysis demonstrates varying degrees of fluconazole susceptibility and distinct Pdr1p mutation profiles, identifying the molecular mechanisms underlying multidrug resistance. Furthermore, the first nosocomial outbreak of Candida auris underscores the importance of multicenter surveillance for identifying and managing C. auris outbreaks. Conclusion: The changing epidemiology of candidemia, along with the continued emergence of antifungal resistance among bloodstream isolates of non-albicans Candida species warrants continuous monitoring of candidemia in Korea. By integrating clinical, microbiological, and public health perspectives, healthcare systems can develop robust strategies to optimize therapeutic approaches, prevent nosocomial transmission, and ultimately reduce morbidity and mortality associated with these life-threatening infections.
Antifungal drug resistance, Candida, Candida auris, Candida glabrata, Candida parapsilosis, Candidemia
The worldwide prevalence of fungal diseases has significantly increased in recent years. Data from more than 120 countries collected between 2010 and 2023 revealed that there are approximately 6.5 million lifethreatening fungal infections annually, with 2.5 million fatalities, which is a disease burden similar to that of tuberculosis, the world’s leading infectious disease [1]. Many fungi cause clinical diseases, Candida and Aspergillus species being the most life threatening [1,2]. Globally, approximately 1,565,000 individuals are affected by candidemia or invasive candidiasis each year, with 995,000 deaths (63.6%) [1]. Candidemia affects at least 600,000 individuals annually, with mortality rates ranging from 30% to 40%, even in highincome countries [2]. Although candidemia is the most common form of invasive candidiasis, culturenegative deep-tissue infections after hematogenous seeding have also been reported [3]. Of the 10,758 Korea Global Antimicrobial Resistance Surveillance System (Kor-GLASS) bloodstream infection (BSI) pathogens identified in 2020–2021, Candida species ranked fourth among all BSI pathogens, followed by Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus. Furthermore, among hospital-origin pathogens, Candida species ranked second, following E. coli. These findings highlight the significant contribution of Candida species in regard to nosocomial BSIs in South Korea [4].
The epidemiology of candidemia has changed over the past few decades. Candida albicans remains the most common isolate in blood cultures; however, infections caused by non-albicans Candida (NAC) species are increasing. The emergence of azole-resistant Candida parapsilosis, multidrug-resistant (MDR) Candida glabrata and Candida auris poses significant global public health challenges [5,6]. In October 2022, the World Health Organization released an initial fungal priority pathogen list that classified 19 fungal species into critical, high, and medium categories, suggesting research priorities [2]. C. auris and C. albicans are of critical concern, while C. glabrata, Candida tropicalis, and C. parapsilosis are of high concern [2]. Candida species differ in their virulence, antifungal susceptibility, and clinical profiles, which must be thoroughly understood if the disease is to be effectively managed [4,7,8].
This review presents an overview of the current epidemiology, antifungal resistance, and clinical implications of Candida species as the primary agents responsible for candidemia. It thoroughly examines recent research conducted in Korea to clarify the reasons for the rising incidence of antifungal drug-resistant pathogens contributing to candidemia in hospitals nationwide. It aims to comprehensively analyze the molecular mechanisms and clinical implications of antifungal resistance in Candida bloodstream isolates, considering the evolving epidemiology of candidemia. We focus on species distribution, antifungal resistance patterns, and clinical outcomes, especially in intensive care units, to identify mechanisms that could improve therapeutic and preventive strategies.
Over 90% of all global candidemia episodes are caused by C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis [4,8,9]. C. albicans is predominant species responsible for candidemia worldwide; however, significant variations exist in the incidence of cases attributed to NAC [4,7,8]. C. glabrata infections are common in both Northern Europe and the United States. In Spain and Brazil, C. parapsilosis infections are more frequent [7]. Early multicenter investigations in South Korea identified C. parapsilosis as the NAC most frequently isolated from candidemia patients [10–12]. However, from 2020 to 2021, C. tropicalis (17.6%) and C. glabrata (17.4%) will be the most common NACs in South Korea [4,8,10–14] (Fig. 1). Similar shifts in the species distribution have been reported in other Korean report [8]. A recent Korean report found that 87.6% of Candida BSI isolates were of hospital origin, and 41.3% were from patients in intensive care units (ICUs) [4]. Adults aged > 60 years accounted for 75.7% of all cases [4]. Candidemia predominantly affects those aged > 60 years; C. glabrata BSIs are most common in the elderly (> 70 years) and C. parapsilosis candidemia is more common in males [4]. The factors driving the evolving epidemiology of NAC in South Korea remain unclear, although certain antifungal medications, infection control measures, and risk profiles of hospitalized patients may play a role [8,9].
A Korean multicenter study evaluated 807 cases of candidemia reported by 11 hospitals in 2017 and 2018. The overall crude 30-day mortality rate was 36.4%. The 30-day mortality rates of patients with candidemia due to C. tropicalis, C. albicans, C. glabrata, and C. parapsilosis were found to be 47.0%, 36.4%, 34.7%, and 22.5%, respectively. Notably, the 30-day mortality rate of ICU-acquired candidemia (ICUAC) was significantly higher than that of non-ICUAC patients (49.5% vs. 25.4 %) for all species (C. albicans, 47.6% vs. 27.7%; C. tropicalis, 57.8% vs. 33.3%; C. glabrata, 50.0% vs. 24.7%; C. parapsilosis, 36.7% vs. 11.3%) [8]. ICU admission was an independent predictor of mortality associated with C. glabrata [odds ratio (OR), 2.07–2.48] and C. parapsilosis (OR, 6.06–11.54) candidemia. Fluconazole resistance predicted C. glabrata associated mortality (OR, 2.80–5.14). An antifungal therapy delay of > 3 days was the strongest predictor of 7-day mortality which was attributable to ICUAC caused by C. albicans (OR, 18.33), C. tropicalis (OR, 10.52), and C. glabrata (OR, 21.30) but was less strongly linked to 30- and 90-day mortality rates (OR, 2.72–6.90). Mortality attributable to C. glabrata ICUAC was found to be more strongly correlated with the absence of antifungal therapy (55.2%) than that attributed to ICUAC caused by other species (30.6%–36.7%). As ICUAC mortality rates and predictors of mortality differ significantly from those in patients with nonICUAC [8], continuous epidemiological surveillance is essential to detect any shift in species distribution and antifungal resistance among Candida BSIs in ICU patients.
The global rise in antifungal-resistant Candida species is particularly concerning, but the rates of resistance to both azoles and echinocandins vary by geographic region, hospital, and ICU. According to the 1997–2016 SENTRY Antimicrobial Surveillance Program, fluconazole resistance in C. glabrata is the highest in North America (10.6%), whereas resistance in C. tropicalis is the highest in the Asia-Pacific region (9.2%) [9]. The prevalence and fluconazole resistance of C. glabrata have increased steadily over the past 20 years in the United States. The echinocandin resistance rates ranged from 3.5% for C. glabrata to 0.1% for C. albicans and C. parapsilosis [9]. The 2019–2020 SENTRY Antimicrobial Surveillance Program reported that 0.5%, 4.5%, 10.5%, and 1.2% of all C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis, respectively, were fluconazole-resistant [15]; among the NAC species, azole resistance was found to be increasing. All C. albicans, C. tropicalis, and Candida krusei isolates, and most C. glabrata (96.2%97.9%) and C. parapsilosis (86.2%–100%) isolates were susceptible to echinocandins [15]. Antifungal resistance rates in bloodstream isolates of the four Candida species from 2005 to 2021 in Korea showed relatively higher fluconazole resistance rates in C. glabrata and C. parapsilosis without consistent trends, whereas echinocandin-resistant isolates were rarely observed (Fig. 2). [4,8,10–14]. The Candida data of KorGLASS obtained from nine sentinel hospitals between 2020 and 2021 revealed that 21.1%, 4.0%, 0.1%, 0.0%, and 0.1% of all Candida isolates in BSIs were not susceptible to fluconazole, voriconazole, caspofungin, micafungin, and anidulafungin, respectively [4]. Fluconazole resistance was apparent in 0% (0/348), 2.2% (3/135), 5.3% (7/133), and 5.6% (6/108) of the C. albicans, C. tropicalis, C. glabrata, and C. parapsilosis BSIs, respectively [4]. The mechanisms of azole antifungal resistance in major Candida species, as identified in Korean multicenter studies, vary among the species (Table 1) [16–20]. Erg11p and Tac1p amino acid substitutions (AASs) may play significant roles in regard to the development of antifungal resistance in C. albicans strains infecting the bloodstream. Most fluconazole-non-susceptible (FNS) infections are not linked to prior azole exposure [16]. In contrast, most FNS C. tropicalis isolates overexpress CDR1, MDR1, and ERG11, and fungemia typically follows azole treatment in immunocompromised patients [17].
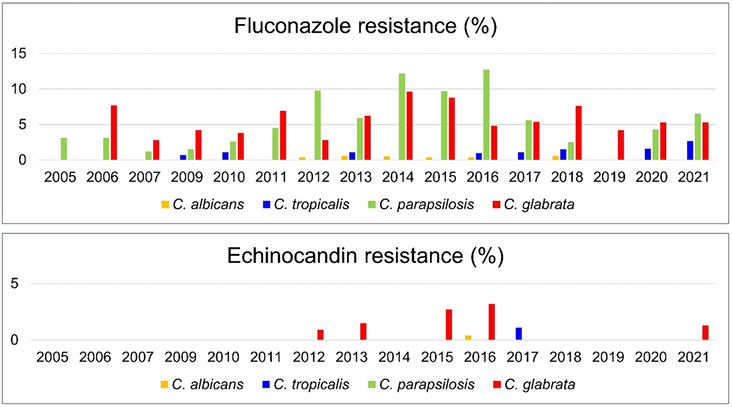
Table 1. Studies describing the azole antifungal resistance mechanisms in the Candida isolates collected from Korean multicenter studies.
| Candida species | C. albicans | C. tropicalis | C. parapsilosis | C. glabrata | C. auris |
|---|---|---|---|---|---|
| Collection period of isolates | 2006–2021 | 2003–2013 | 2005–2016 | 2008–2018 | 1996–2022 |
| Number of hospitals | 10 | 8 | 8 | 19 | 13 |
| Number of isolates | 26 BSI (14 FNS and 12 FS) isolates | 9 FNS, 12 FS (MIC, 1–2 µg/ml), and 14 control (MIC, 0.125–0.5 µg/ml) isolates | 67 BSI (47 FR and 20 FS) isolates | 278 BSI (66 FR and 212 F-SDD) isolates | 104 (96 clade II, and 8 clade I) isolates |
| Genes studied | Sequencing of ERG11, TAC1, MRR1, and UPC2 | Quantitation of CDR1, MDR1, and ERG11 expression, and sequencing of the ERG11 and UPC2 genes | Sequencing of ERG11, TAC1, MRR1, and UPC2 | Sequencing of pleiotropic drug resistance transcription factor (PDR1) | Sequencing of ERG11, TAC1A, and TAC1B |
| Results | Erg11p and Tac1p AASs are likely to contribute to FR in C. albicans BSI isolates in Korea | The majority of FNS C. tropicalis isolates show overexpression of CDR1, MDR1, and ERG11 genes | Majority (63.8%) of the C. parapsilosis FR isolates exhibit the Y132F substitution in Erg11p | Most FR BSI isolates of C. glabrata in Korea harbor FR-specific Pdr1p AAS | Tac1Bp AASs may be the predominant fluconazole resistance mechanism in clade II Korean isolates of C. auris |
| Year of publication | 2023 | 2016 | 2018 | 2021 | 2023 |
| References | [16] | [17] | [18] | [19] | [20] |
Abbreviations: BSI, bloodstream infection; FNS, fluconazole non-susceptible; FS, fluconazole susceptible; F-SDD, fluconazole susceptible dose-dependent; FR, fluconazole resistant; MIC, minimal inhibitory concentration; AAS, amino acid substitution.
Candida species, predominantly C. albicans and C. tropicalis (with lower proportions of C. parapsilosis and C. glabrata), are integral components of the normal gastrointestinal microbiome [21]. Most candidemia cases are endogenous infections, C. albicans (which constitutes 70%–80% of commensal Candida) is the most common causative agent [22]. The main mechanism of candidemia is thought to be the leakage of Candida through the intestinal mucosa following mucosal insults [23], such as those caused by abdominal surgery, anastomotic leaks, antibiotic use, and pancreatitis. In addition, Candida often gains bloodstream access via intravenous catheters, particularly in central venous lines. In patients receiving total parenteral nutrition (TPN), lipid emulsions enhance Candida biofilm formation, potentially increasing its virulence. Altered gut function in TPN recipients may also be conducive for Candida translocation [24]. Many molecular typing studies have shown that small clusters of candidemia attributable to isogenic isolates are frequent within hospitals, Candida can spread nosocomially [18,25,26] via the hands of healthcare personnel or contamination of intravenous saline used to flush central venous catheters (CVCs) shared among patients. In addition, personto-person transmission among hospitalized patients may have occurred more frequently than previously thought. The clonal spread of C. albicans causing candidemia is common in hospitals [25,26]. Outbreaks of C. albicans often occur over a short period and simultaneously involve multiple patients. In contrast, C. parapsilosis is more persistent in healthcare settings, with clusters of infected patients developing over time [27], likely due to its frequent presence on the hands of healthy individuals and healthcare workers [6]. Strict adherence to routine hand hygiene and the appropriate management of CVCs are of utmost importance [6]. “Catheter care bundles,” which are standardized interventions that prevent catheter-associated BSIs, significantly decrease the incidence of candidemia [6].
C. parapsilosis is the second most common cause of Candida BSIs in Latin America, Asia, and Southern Europe, and the third most common global BSI cause [5,28]. C. parapsilosis candidemia is frequently attributed to external fungal acquisition; C. parapsilosis tends to colonize hospital settings and equipment, particularly CVCs and other medical devices [29]. One Korean study explored whether biofilm formation was associated with clinical features [30]. The proportion of blood NAC isolates that formed biofilms was significantly higher than that of NAC isolates from other locations (79% vs. 52%). Specifically, bloodstream isolates of C. parapsilosis demonstrated a significantly higher likelihood of being biofilm positive than those obtained from other sites (86% vs. 47%). All NAC species, including C. parapsilosis, were more likely to form biofilms when isolated from patients with CVC-related candidemia associated with TPN than from other patients. The ability of in vitro biofilm formation in C. parapsilosis when grown in glucose-containing Sabouraud dextrose broth is an important virulence factor that causes CVC-related fungemia in patients treated with TPN [30].
Although C. parapsilosis is usually susceptible to azole antifungals, C. parapsilosis BSIs caused by FNS isolates have become increasingly common worldwide [5,26]. The global prevalence of fluconazole resistance ranges from 0% to 100%, and the pooled resistance rate may reach 15.2% [31]. Recent reports indicated that 60.7%-63.8% FNS C. parapsilosis isolates harbored a Y132F substitution in the ERG11 gene (“Y132F isolates”), and most were clonally related. BSI outbreaks caused by Y132F isolates have been reported in multiple countries across four continents, and some clonal isolates have become endemic over several years in affected hospitals [18,32]. An early multicenter Korean report found that among 1,009 nonduplicated BSIs of C. parapsilosis obtained from 20 hospitals between 2005 and 2016, 47 (4.7%) from eight university hospitals were fluconazole resistant and 64% (30/47) harbored the same Y132F mutation in Erg11p [18].
A recent Korean study found that long-term clonal transmission of C. parapsilosis BSI Y132F isolates in Korean hospitals was associated with a “sinking” (not a “floating”) phenotype [33]. In contrast to floating phenotypes, sinking phenotypes are presented as a smaller, button-like appearance in the plate well of Clinical and Laboratory Standards Institute broth microdilution antifungal susceptibility test; all cells sank in U-shaped, round-bottom wells. The sinking phenotype was detected in 86.7% of the FNS BSI isolates, and 92.9% of the Y132F BSI isolates of C. parapsilosis. As azole breakthrough fungemia, ICU admission, and urinary catheter use were independent risk factors for fungemia caused by Y132F sinking phenotype isolates, strain with a sinking phenotype may be prone to having an ERG11 Y132F substitution after azole exposure, may be more enriched, and may persist for a long time in the ICU environment, where they may cause BSI in vulnerable patients with indwelling catheters or azole exposure [33]. Microsatellite typing revealed that these isolates exhibited greater clonal transmission than other fluconazole-resistant isolates and persisted in hospitals for several years. The evolution of fluconazole resistance in Y132F clonal C. parapsilosis isolates from a South Korean hospital has been studied [34]. Increased fluconazole resistance has been associated with the acquisition of MRR1 mutations. Thus, continuous surveillance of fluconazole resistance rates, resistance mechanism(s), and clonality of hospital C. parapsilosis isolates is essential [33,34].
C. glabrata is one of the most concerning Candida species of nosocomial importance; its antifungal drug resistance rate is increasing, and MDR isolates are emerging [4,9,35]. C. glabrata is innately relatively resistant to azoles, especially fluconazole, and can rapidly develop fluconazole resistance during treatment, probably because its haploid genome is mutable. C. glabrata isolates are no longer considered fluconazolesusceptible but rather fluconazole-susceptible dose-dependent (F-SDD) or fluconazole-resistant [35,36]. Although the incidences of echinocandin-resistant and MDR C. glabrata BSIs are low, fluconazole-resistant C. glabrata BSIs have been increasingly reported worldwide, typically at a rate of 2.6%–10.6% but increasing up to 17% [9].
Gain-of-function (GOF) mutations in the transcription factor pleiotropic drug resistance protein 1 (encoded by PDR1) mediate C. glabrata azole resistance by controlling the expression of genes encoding efflux pumps [CDR1, CDR2 (PDH1), and SNQ2], although other mechanisms may also be involved [37]. A Korean multicenter study found that 98.5% of fluconazole-resistant C. glabrata BSIs and 0.9% of C. glabrata F-SDD BSIs exhibited one or two AASs in the Pdr1p protein, excluding five genotype-specific AASs [19]. Of the 49 Pdr1p AASs, 33 were found to be new; most fluconazole-resistant BSIs harbored fluconazole resistancespecific Pdr1p AASs. More importantly, patients infected with fluconazole-resistant C. glabrata BSIs harboring Pdr1p mutations exhibited high mortality; the 30-day rate for 64 patients was 60.9%, and the 90day rate was 78.2%, which was significantly higher than that for patients with F-SDD BSIs (30-day rate, 36.4%; 90-day rate, 43.8%) [19].
The echinocandin resistance rate of C. glabrata ranged from 1.7% to 3.5% and resistance did not increase over time [9,38]. However, at the institutional level, the prevalence of echinocandin resistance varies significantly, and sometimes exceeds 13% [9,38]. A recent Korean study used whole-genome sequencing (WGS) in order to explore the molecular mechanisms of MDR in 10 serial C. glabrata isolates from a patient with BSI experiencing breakthrough fungemia during extended amphotericin B (AMB)/echinocandin therapy [39]. ERG3 and ERG6 may be involved in AMB resistance, and Fks2p mutations outside the highsusceptibility regions contribute to low echinocandin resistance [39]. The minimal inhibitory concentration (MIC) of fluconazole for C. glabrata isolates with the same Pdr1p GOF mutation was reduced in AMBresistant isolates with Erg6p mutations, indicating a complex relationship between AMB and azole resistance. Changes in the ergosterol biosynthesis pathway may explain the azole resistance in some C. glabrata strains [39].
Although C. glabrata transmission within hospitals is less common than that of other Candida species, some transmission has been observed [40,41]. The prevalence of antifungal-resistant BSIs caused by C. glabrata is increasing worldwide [5,9,19], but the transmission of such resistant strains within hospitals has rarely been reported. However, the Centers for Disease Control and Prevention’s Emerging Infections Program in USA found an exceptionally high proportion of echinocandin-resistant C. glabrata with the FKS mutations seen among echinocandin-naïve patients, suggesting nosocomial transmission [42]. Using WGS and epidemiological analyses, a recent Korean study identified two potential clusters of C. glabrata BSIs within the same hospital. Clonal C. glabrata strains with differences in fluconazole susceptibility and distinct Pdr1p AAS profiles were isolated from patients receiving antifungal therapy [43].
C. auris poses a significant threat to global health [2,44]. C. auris was first described in Japan and South Korea in 2009 [45,46]. The first Japanese C. auris was isolated from an inpatient’s external ear canal [45]. In the same year, Candida haemulonii and other closely related species from five Korean university hospitals were reported [46]. Of these, 15 ear isolates of the new Candida species were identified as C. auris; all from patients with chronic otitis media. Persistently positive cultures were obtained from seven patients, three of whom received antifungal therapy. However, no histopathological evidence of a fungal infection was observed in any patient. Thus, the clinical significance of C. auris isolation from the ears remains unclear. Two years later, the first three cases of C. auris fungemia were identified at three Korean university hospitals [47]. The first case was incidentally discovered through molecular analysis of an unidentified yeast isolated in 1996; the earliest strain of C. auris was thus from Korea. Two of these three cases were identified in the 2009 Korean multicenter candidemia surveillance study. These three cases show, for the first time, that C. auris is a human pathogen. Since then, C. auris has been found in more than 45 countries across six continents [48] and has been associated with many outbreaks of invasive diseases. The mortality rate ranged from 29% to 62%, and the median hospital stay was typically 46–68 days but reached up to 140 days [49].
C. auris differs from other Candida species in three respects [50]. First, C. auris is primarily associated with the skin and not the gut or mucosal surfaces. Second, because C. auris can persist on the skin, it can spread widely in healthcare environments, affecting both patients and the environment, and hospital outbreaks are frequent. Third, C. auris is the first nosocomial fungal pathogen to exhibit marked (sometimes complete) resistance to all known antifungals, including azoles, AMB, and echinocandins. The global fluconazole resistance rate was 87%–100%. The MIC90 of isavuconazole, itraconazole, and posaconazole are 0.06–1.0 mg/L. The voriconazole resistance rate was 28%–98%. The AMB resistance rate is 8%–35% and that of echinocandin resistance is 0%–8% [49].
In the initial WGS analysis, C. auris isolates were grouped into four major clades: South Asia clade I, East Asia clade II, African clade III, and South American clade IV [51]. Only clade II isolates were detected in Korean hospitals between 1996 and 2018 [52]. Pulsed-field gel electrophoresis showed that the patterns of the Korean blood and ear isolates were similar or identical and distinct from those of other countries. Almost all Korean isolates were obtained from ears. No nosocomial outbreaks caused by clade II C. auris have yet been reported. Two Korean studies found that 66% of 157 isolates of clade II C. auris were fluconazoleresistant, but all were susceptible to AMB and the three echinocandins, and MDR was not found [20,52]. Thus, clade II Korean isolates of C. auris are less resistant to antifungals and differ in terms of clinical characteristics compared to other geographic clades [52]. In 2022, Kor-GLASS identified nine C. auris clade I BSIs in a single hospital [20]. This is the first nosocomial outbreak in Korea, highlighting the importance of multicenter surveillance for identifying and managing C. auris outbreaks. Clade II Korean isolates were less tolerant to 42°C than clade I isolates. Furthermore, in a model using Galleria mellonella larvae, the clade II isolates were less virulent than the clade I isolates [20]. The growth rate of clade II isolates was fivefold lower than that of C. albicans SC5314, but the clade I growth rate was 4.3-fold higher than that of C. albicans. The low virulence of clade II may explain its lack of nosocomial transmission [20]. A detailed summary of microbiological studies on the clinical isolates of C. auris in Korea is provided in Table 2. Infections caused by C. auris have been documented in at least 40 countries across six continents. Although clade II C. auris lacks MDR and does not appear to cause nosocomial candidemia, the increasing incidence of clade I C. auris infections in Korea is alarming. The timely identification and implementation of effective control measures are critical.
Table 2. Summary of the microbiological studies of the clinical isolates of Candida auris in Korea
| Year of publication | No. of patient (No. of isolate) | Specimen (No. of isolate) | Molecular epidemiologic test (No. of isolate)a | Resistance rate of antifungal agent (%)b | Possible genetic marker of FLU resistant isolate | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| FLU | AMB | CSF | MCF | ANF | ||||||
| 2009 | 15 (15) | Ear (15) | NT | 53.3 | 0 | 0 | 0 | NT | NT | [46] |
| 2011 | 3 (6) | Blood (6) | NT | 33.3 | 0 | 0 | 0 | NT | NT | [44] |
| 2018 | 61 (61) | Blood (4), Ear (57) | Clade II (61) | 62.3 | 0 | 0 | 0 | NT | Erg11p (L43H, K143R, Q357K) | [52] |
| 2023 | 104 (104) | Blood (5), Ear (91) | Clade I (8) | 25.0 | 75.0 | 0 | 0 | 0 | Erg11p (K143R), Tac1Bp (A640V) | [20] |
| Clade II (96) | 68.8 | 0 | 0 | 0 | 0 | Erg11p (L43H, Y132F, K143R, Q357K), Tac1Bp (F214S, P595L) | ||||
aClade of isolates were determined by multilocus sequence typing.
bApplying tentative breakpoints of the Centers for Disease Control and Prevention.
Abbreviations: FLU, fluconazole; AMB, amphotericin B; CSF, Caspofungin; MCF, micafungin; ANF, anidulafungin; NT, not tested.
Candidemia, the epidemiology of which differs geographically, remains an important issue. The global landscape has evolved over time and is likely influenced by selective drug pressure, innate characteristics of Candida species, and host- and drug-related parameters. Increasing antifungal resistance, particularly in azoleresistant C. parapsilosis, MDR C. glabrata and C. auris, complicates treatment strategies and is associated with an increasing incidence of candidemia in healthcare settings. The interrelationships between virulence, epidemiology, and antifungal susceptibility/resistance must be understood if management is effective. Because person-to-person transmission among hospitalized patients may be more frequent than previously recognized, adherence to established infection prevention protocols is imperative. WGS will yield insights into drug resistance mechanisms, virulence factors, genotypic characteristics, and sources of infection. Given the link between antifungal resistance in Candida isolates and their evolutionary adaptations, it is vital to sustain the ongoing multicenter surveillance of antifungal resistance rates, the underlying mechanisms of resistance, and also the clonal relationships of Candida isolates sourced from healthcare settings.
This was not a human population study; therefore, approval by the institutional review board and informed consent were not required.
No potential conflicts of interest relevant to this article were reported.
This study was supported by a Research Program funded by the Korea Centers for Disease Control and Prevention (grant 2023-10-004).
None.
1. Denning DW. Global incidence and mortality of severe fungal disease. Lancet Infect Dis 2024;24:e428-38.

2. Casalini G, Giacomelli A, Antinori S. The WHO fungal priority pathogens list: a crucial reappraisal to review the prioritisation. Lancet Microbe 2024;5:717-24.

3. Kullberg BJ and Arendrup MC. Invasive candidiasis. N Engl J Med 2015;373:1445-56.

4. Won EJ, Choi MJ, Jeong SH, Kim D, Shin KS, Shin JH, et al. Nationwide surveillance of antifungal resistance of Candida bloodstream isolates in South Korean hospitals: two year report from Kor-GLASS. J Fungi 2022;8:996.


5. Arastehfar A, Lass-Flörl C, Garcia-Rubio R, Daneshnia F, Ilkit M, Boekhout T, et al. The quiet and underappreciated rise of drug-resistant invasive fungal pathogens. J Fungi 2020;6:138.


6. Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. Invasive candidiasis. Nat Rev Dis Primers 2018;4:18026.

7. Guinea J. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 2014;20(Suppl 6):5-10.
8. Kwon YJ, Won EJ, Jeong SH, Shin KS, Shin JH, Kim YR, et al. Dynamics and predictors of mortality due to candidemia caused by different Candida species: comparison of intensive care unit-associated candidemia (ICUAC) and Non-ICUAC. J Fungi 2021;7:597.


9. Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997-2016. Open Forum Infect Dis 2019;6:S79-94.


10. Won EJ, Shin JH, Choi MJ, Lee WG, Park YJ, Uh Y, et al. Antifungal susceptibilities of bloodstream isolates of Candida species from nine hospitals in Korea: application of new antifungal breakpoints and relationship to antifungal usage. PLoS One 2015;10:e0118770.


11. Jung SI, Shin JH, Song JH, Peck KR, Lee K, Kim MN, et al. Multicenter surveillance of species distribution and antifungal susceptibilities of Candida bloodstream isolates in South Korea. Med Mycol 2010;48:669-74.

12. Lee JS, Shin JH, Lee K, Kim MN, Shin BM, Uh Y, et al. Species distribution and susceptibility to azole antifungals of Candida bloodstream isolates from eight university hospitals in Korea. Yonsei Med J 2007;48:779-86.


13. Jung SI, Shin JH, Choi HJ, Ju MY, Kim SH, Lee WG, et al. Antifungal susceptibility to amphotericin B, fluconazole, voriconazole, and flucytosine in Candida bloodstream isolates from 15 tertiary hospitals in Korea. Ann Lab Med 2012;32:426-8.


14. Jang MJ, Shin JH, Lee WG, Kim MN, Lee K, Lee HS, et al. In vitro fluconazole and voriconazole susceptibilities of Candida bloodstream isolates in Korea: use of the CLSI and EUCAST epidemiological cutoff values. Ann Lab Med 2013;33:167-73.


15. Carvalhaes CG, Klauer AL, Rhomberg PR, Pfaller MA, Castanheira M. Evaluation of rezafungin provisional CLSI clinical breakpoints and epidemiological cutoff values tested against a worldwide collection of contemporaneous invasive fungal isolates (2019 to 2020). J Clin Microbiol 2022;60:e0244921.


16. Choi MJ, Kwon YJ, Byun SA, Kim MN, Lee WG, Lee J, et al. Molecular and clinical features of fluconazole non-susceptible Candida albicans bloodstream isolates recovered in Korean multicenter surveillance studies. Ann Lab Med 2023;43:614-9.


17. Choi MJ, Won EJ, Shin JH, Kim SH, Lee WG, Kim MN, et al. Resistance mechanisms and clinical features of fluconazole-nonsusceptible Candida tropicalis isolates compared with f luconazole-less-susceptible isolates. Antimicrob Agents Chemother 2016;60:3653-61.


18. Choi YJ, Kim YJ, Yong D, Byun JH, Kim TS, Chang YS, et al. Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F mutation in ERG11 gene, South Korea. Emerg Infect Dis 2018;24:1768-70.


19. Won EJ, Choi MJ, Kim MN, Yong D, Lee WG, Uh Y, et al. Fluconazole-resistant Candida glabrata bloodstream isolates, South Korea, 2008-2018. Emerg Infect Dis 2021;27:779-88.


20. Byun SA, Kwon YJ, Lee GY, Choi MJ, Jeong SH, Kim D, et al. Virulence traits and azole resistance in Korean Candida auris isolates. J Fungi 2023;9:979.


21. Li J, Chen D, Yu B, He J, Zheng P, Mao X, et al. Fungi in gastrointestinal tracts of human and mice: from community to functions. Microb Ecol 2018;75:821-9.

22. Chin VK, Lee TY, Rusliza B, Chong PP. Dissecting Candida albicans infection from the perspective of C. albicans virulence and omics approaches on host-pathogen interaction: a review. Int J Mol Sci 2016;17:1643.


23. Mora Carpio AL and Climaco A. Candidemia. [Online] (last visited on 8 August 2023).
24. Swindell K, Lattif AA, Chandra J, Mukherjee PK, Ghannoum MA. Parenteral lipid emulsion induces germination of Candida albicans and increases biofilm formation on medical catheter surfaces. J Infect Dis 2009;200:473-80.

25. Shin JH, Park MR, Song JW, Shin DH, Jung SI, Cho D, et al. Microevolution of Candida albicans strains during catheter-related candidemia. J Clin Microbiol 2004;42:4025-31.


26. Escribano P, Rodríguez-Créixems M, Sánchez-Carrillo C, Muñoz P, Bouza E, Guinea J. Endemic genotypes of Candida albicans causing fungemia are frequent in the hospital. J Clin Microbiol 2013;51:2118-23.


27. Guinea J, Mezquita S, Gómez A, Padilla B, Zamora E, Sánchez-Luna M, et al. Whole genome sequencing confirms Candida albicans and Candida parapsilosis microsatellite sporadic and persistent clones causing outbreaks of candidemia in neonates. Med Mycol 2021;60:myab068.

28. Pfaller MA, Jones RN, Doern GV, Sader HS, Messer SA, Houston A, et al. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997-1998. Antimicrob Agents Chemother 2000;44:747-51.


29. Trofa D, Gácser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev 2008;21:606-25.


30. Shin JH, Kee SJ, Shin MG, Kim SH, Shin DH, Lee SK, et al. Biofilm production by isolates of Candida species recovered from nonneutropenic patients: comparison of bloodstream isolates with isolates from other sources. J Clin Microbiol 2002;40:1244-8.


31. Yamin D, Akanmu MH, Al Mutair A, Alhumaid S, Rabaan AA, Hajissa K. Global prevalence of antifungal-resistant Candida parapsilosis: a systematic review and meta-analysis. Trop Med Infect Dis 2022;7:188.


32. Arastehfar A, Hilmioğlu-Polat S, Daneshnia F, Pan W, Hafez A, Fang W, et al. Clonal candidemia outbreak by Candida parapsilosis carrying Y132F in Turkey: evolution of a persisting challenge. Front Cell Infect Microbiol 2021;11:676177.


33. Byun JH, Won EJ, Cho HW, Kim D, Lee H, Kim SH, et al. Detection and characterization of two phenotypes of Candida parapsilosis in South Korea: clinical features and microbiological findings. Microbiol Spectr 2023;11:e0006623.


34. Kim TY, Huh HJ, Lee GY, Choi MJ, Yu HJ, Cho SY, et al. Evolution of fluconazole resistance mechanisms and clonal types of Candida parapsilosis isolates from a tertiary care hospital in South Korea. Antimicrob Agents Chemother 2022;66:e0088922.


35. Arendrup MC and Patterson TF. Multidrug-resistant Candida: epidemiology, molecular mechanisms, and treatment. J Infect Dis 2017;216(suppl_3):S445-51.

36. Clinical and Laboratory Standard Institute. Performance standards for antifungal susceptibility testing of yeasts. Document M60Ed2. 2nd ed. Wayne, PA; CLSI: 2020.
37. Ferrari S, Ischer F, Calabrese D, Posteraro B, Sanguinetti M, Fadda G, et al. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog 2009;5:e1000268.


38. Bassetti M, Vena A, Bouza E, Peghin M, Muñoz P, Righi E, et al. Antifungal susceptibility testing in Candida, Aspergillus and Cryptococcus infections: are the MICs useful for clinicians? Clin Microbiol Infect 2020;26:1024-33.

39. Lim HJ, Choi MJ, Byun SA, Won EJ, Park JH, Choi YJ, et al. Whole-genome sequence analysis of Candida glabrata isolates from a patient with persistent fungemia and determination of the molecular mechanisms of multidrug resistance. J Fungi 2023;9:515.


40. Goemaere B, Lagrou K, Spriet I, Hendrickx M, Becker P. Clonal spread of Candida glabrata bloodstream isolates and fluconazole resistance affected by prolonged exposure: a 12-year single-center study in Belgium. Antimicrob Agents Chemother 2018;62:e00591-18.


41. Katiyar S, Shiffrin E, Shelton C, Healey K, Vermitsky JP, Edlind T. Evaluation of polymorphic locus sequence typing for Candida glabrata epidemiology. J Clin Microbiol 2016;54:1042-50.


42. Vallabhaneni S, Cleveland AA, Farley MM, Harrison LH, Schaffner W, Beldavs ZG, et al. Epidemiology and risk factors for echinocandin nonsusceptible Candida glabrata bloodstream infections: data from a large multisite population-based candidemia surveillance program, 2008-2014. Open Forum Infect Dis 2015;2:ofv163.


43. Hwang IJ, Kwon YJ, Lim HJ, Hong KH, Lee H, Yong D, et al. Nosocomial transmission of fluconazole-resistant Candida glabrata bloodstream isolates revealed by whole-genome sequencing. Microbiol Spectr 2024;e0088324.


44. U.S. Centers for Disease Control and Prevention. 2019 Antibiotic resistance threats report. [Online] (last visited on 4 December 2024).
45. Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 2009;53:41-4.

46. Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 2009;48:e57-61.

47. Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, et al. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 2011;49:3139-42.


48. Wang Y and Xu J. Associations between genomic variants and antifungal susceptibilities in the archived global Candida auris population. J Fungi 2024;10:86.


49. Kim HY, Nguyen TA, Kidd S, Chambers J, Alastruey-Izquierdo A, Shin JH, et al. Candida auris-a systematic review to inform the world health organization fungal priority pathogens list. Med Mycol 2024;62:myae042.


50. Lockhart SR, Chowdhary A, Gold JAW. The rapid emergence of antifungal-resistant human-pathogenic fungi. Nat Rev Microbiol 2023;21:818-32.


51. Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 2017;64:134-40.


52. Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, et al. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol 2019;57:e01624-18.


1. Denning DW. Global incidence and mortality of severe fungal disease. Lancet Infect Dis 2024;24:e428-38.
2. Casalini G, Giacomelli A, Antinori S. The WHO fungal priority pathogens list: a crucial reappraisal to review the prioritisation. Lancet Microbe 2024;5:717-24.
3. Kullberg BJ and Arendrup MC. Invasive candidiasis. N Engl J Med 2015;373:1445-56.
4. Won EJ, Choi MJ, Jeong SH, Kim D, Shin KS, Shin JH, et al. Nationwide surveillance of antifungal resistance of Candida bloodstream isolates in South Korean hospitals: two year report from Kor-GLASS. J Fungi 2022;8:996.
5. Arastehfar A, Lass-Flörl C, Garcia-Rubio R, Daneshnia F, Ilkit M, Boekhout T, et al. The quiet and underappreciated rise of drug-resistant invasive fungal pathogens. J Fungi 2020;6:138.
6. Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. Invasive candidiasis. Nat Rev Dis Primers 2018;4:18026.
7. Guinea J. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 2014;20(Suppl 6):5-10.
8. Kwon YJ, Won EJ, Jeong SH, Shin KS, Shin JH, Kim YR, et al. Dynamics and predictors of mortality due to candidemia caused by different Candida species: comparison of intensive care unit-associated candidemia (ICUAC) and Non-ICUAC. J Fungi 2021;7:597.
9. Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997-2016. Open Forum Infect Dis 2019;6:S79-94.
10. Won EJ, Shin JH, Choi MJ, Lee WG, Park YJ, Uh Y, et al. Antifungal susceptibilities of bloodstream isolates of Candida species from nine hospitals in Korea: application of new antifungal breakpoints and relationship to antifungal usage. PLoS One 2015;10:e0118770.
11. Jung SI, Shin JH, Song JH, Peck KR, Lee K, Kim MN, et al. Multicenter surveillance of species distribution and antifungal susceptibilities of Candida bloodstream isolates in South Korea. Med Mycol 2010;48:669-74.
12. Lee JS, Shin JH, Lee K, Kim MN, Shin BM, Uh Y, et al. Species distribution and susceptibility to azole antifungals of Candida bloodstream isolates from eight university hospitals in Korea. Yonsei Med J 2007;48:779-86.
13. Jung SI, Shin JH, Choi HJ, Ju MY, Kim SH, Lee WG, et al. Antifungal susceptibility to amphotericin B, fluconazole, voriconazole, and flucytosine in Candida bloodstream isolates from 15 tertiary hospitals in Korea. Ann Lab Med 2012;32:426-8.
14. Jang MJ, Shin JH, Lee WG, Kim MN, Lee K, Lee HS, et al. In vitro fluconazole and voriconazole susceptibilities of Candida bloodstream isolates in Korea: use of the CLSI and EUCAST epidemiological cutoff values. Ann Lab Med 2013;33:167-73.
15. Carvalhaes CG, Klauer AL, Rhomberg PR, Pfaller MA, Castanheira M. Evaluation of rezafungin provisional CLSI clinical breakpoints and epidemiological cutoff values tested against a worldwide collection of contemporaneous invasive fungal isolates (2019 to 2020). J Clin Microbiol 2022;60:e0244921.
16. Choi MJ, Kwon YJ, Byun SA, Kim MN, Lee WG, Lee J, et al. Molecular and clinical features of fluconazole non-susceptible Candida albicans bloodstream isolates recovered in Korean multicenter surveillance studies. Ann Lab Med 2023;43:614-9.
17. Choi MJ, Won EJ, Shin JH, Kim SH, Lee WG, Kim MN, et al. Resistance mechanisms and clinical features of fluconazole-nonsusceptible Candida tropicalis isolates compared with f luconazole-less-susceptible isolates. Antimicrob Agents Chemother 2016;60:3653-61.
18. Choi YJ, Kim YJ, Yong D, Byun JH, Kim TS, Chang YS, et al. Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F mutation in ERG11 gene, South Korea. Emerg Infect Dis 2018;24:1768-70.
19. Won EJ, Choi MJ, Kim MN, Yong D, Lee WG, Uh Y, et al. Fluconazole-resistant Candida glabrata bloodstream isolates, South Korea, 2008-2018. Emerg Infect Dis 2021;27:779-88.
20. Byun SA, Kwon YJ, Lee GY, Choi MJ, Jeong SH, Kim D, et al. Virulence traits and azole resistance in Korean Candida auris isolates. J Fungi 2023;9:979.
21. Li J, Chen D, Yu B, He J, Zheng P, Mao X, et al. Fungi in gastrointestinal tracts of human and mice: from community to functions. Microb Ecol 2018;75:821-9.
22. Chin VK, Lee TY, Rusliza B, Chong PP. Dissecting Candida albicans infection from the perspective of C. albicans virulence and omics approaches on host-pathogen interaction: a review. Int J Mol Sci 2016;17:1643.
23. Mora Carpio AL and Climaco A. Candidemia. [Online] (last visited on 8 August 2023).
24. Swindell K, Lattif AA, Chandra J, Mukherjee PK, Ghannoum MA. Parenteral lipid emulsion induces germination of Candida albicans and increases biofilm formation on medical catheter surfaces. J Infect Dis 2009;200:473-80.
25. Shin JH, Park MR, Song JW, Shin DH, Jung SI, Cho D, et al. Microevolution of Candida albicans strains during catheter-related candidemia. J Clin Microbiol 2004;42:4025-31.
26. Escribano P, Rodríguez-Créixems M, Sánchez-Carrillo C, Muñoz P, Bouza E, Guinea J. Endemic genotypes of Candida albicans causing fungemia are frequent in the hospital. J Clin Microbiol 2013;51:2118-23.
27. Guinea J, Mezquita S, Gómez A, Padilla B, Zamora E, Sánchez-Luna M, et al. Whole genome sequencing confirms Candida albicans and Candida parapsilosis microsatellite sporadic and persistent clones causing outbreaks of candidemia in neonates. Med Mycol 2021;60:myab068.
28. Pfaller MA, Jones RN, Doern GV, Sader HS, Messer SA, Houston A, et al. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997-1998. Antimicrob Agents Chemother 2000;44:747-51.
29. Trofa D, Gácser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev 2008;21:606-25.
30. Shin JH, Kee SJ, Shin MG, Kim SH, Shin DH, Lee SK, et al. Biofilm production by isolates of Candida species recovered from nonneutropenic patients: comparison of bloodstream isolates with isolates from other sources. J Clin Microbiol 2002;40:1244-8.
31. Yamin D, Akanmu MH, Al Mutair A, Alhumaid S, Rabaan AA, Hajissa K. Global prevalence of antifungal-resistant Candida parapsilosis: a systematic review and meta-analysis. Trop Med Infect Dis 2022;7:188.
32. Arastehfar A, Hilmioğlu-Polat S, Daneshnia F, Pan W, Hafez A, Fang W, et al. Clonal candidemia outbreak by Candida parapsilosis carrying Y132F in Turkey: evolution of a persisting challenge. Front Cell Infect Microbiol 2021;11:676177.
33. Byun JH, Won EJ, Cho HW, Kim D, Lee H, Kim SH, et al. Detection and characterization of two phenotypes of Candida parapsilosis in South Korea: clinical features and microbiological f indings. Microbiol Spectr 2023;11:e0006623.
34. Kim TY, Huh HJ, Lee GY, Choi MJ, Yu HJ, Cho SY, et al. Evolution of fluconazole resistance mechanisms and clonal types of Candida parapsilosis isolates from a tertiary care hospital in South Korea. Antimicrob Agents Chemother 2022;66:e0088922.
35. Arendrup MC and Patterson TF. Multidrug-resistant Candida: epidemiology, molecular mechanisms, and treatment. J Infect Dis 2017;216(suppl_3):S445-51.
36. Clinical and Laboratory Standard Institute. Performance standards for antifungal susceptibility testing of yeasts. Document M60Ed2. 2nd ed. Wayne, PA; CLSI: 2020.
37. Ferrari S, Ischer F, Calabrese D, Posteraro B, Sanguinetti M, Fadda G, et al. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog 2009;5:e1000268.
38. Bassetti M, Vena A, Bouza E, Peghin M, Muñoz P, Righi E, et al. Antifungal susceptibility testing in Candida, Aspergillus and Cryptococcus infections: are the MICs useful for clinicians? Clin Microbiol Infect 2020;26:1024-33.
39. Lim HJ, Choi MJ, Byun SA, Won EJ, Park JH, Choi YJ, et al. Whole-genome sequence analysis of Candida glabrata isolates from a patient with persistent fungemia and determination of the molecular mechanisms of multidrug resistance. J Fungi 2023;9:515.
40. Goemaere B, Lagrou K, Spriet I, Hendrickx M, Becker P. Clonal spread of Candida glabrata bloodstream isolates and fluconazole resistance affected by prolonged exposure: a 12-year single-center study in Belgium. Antimicrob Agents Chemother 2018;62:e00591-18.
41. Katiyar S, Shiffrin E, Shelton C, Healey K, Vermitsky JP, Edlind T. Evaluation of polymorphic locus sequence typing for Candida glabrata epidemiology. J Clin Microbiol 2016;54:1042-50.
42. Vallabhaneni S, Cleveland AA, Farley MM, Harrison LH, Schaffner W, Beldavs ZG, et al. Epidemiology and risk factors for echinocandin nonsusceptible Candida glabrata bloodstream infections: data from a large multisite population-based candidemia surveillance program, 2008-2014. Open Forum Infect Dis 2015;2:ofv163.
43. Hwang IJ, Kwon YJ, Lim HJ, Hong KH, Lee H, Yong D, et al. Nosocomial transmission of fluconazole-resistant Candida glabrata bloodstream isolates revealed by whole-genome sequencing. Microbiol Spectr 2024:e0088324.
44. U.S. Centers for Disease Control and Prevention. 2019 Antibiotic resistance threats report. [Online] (last visited on 4 December 2024).
45. Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 2009;53:41-4.
46. Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 2009;48:e57-61.
47. Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, et al. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 2011;49:3139-42.
48. Wang Y and Xu J. Associations between genomic variants and antifungal susceptibilities in the archived global Candida auris population. J Fungi 2024;10:86.
49. Kim HY, Nguyen TA, Kidd S, Chambers J, Alastruey-Izquierdo A, Shin JH, et al. Candida auris-a systematic review to inform the world health organization fungal priority pathogens list. Med Mycol 2024;62:myae042.
50. Lockhart SR, Chowdhary A, Gold JAW. The rapid emergence of antifungal-resistant humanpathogenic fungi. Nat Rev Microbiol 2023;21:818-32.
51. Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 2017;64:134-40.
52. Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, et al. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol 2019;57:e01624-18.