Jaesoon Kim, Nakwon Kwak, Jee-Soo Lee, Taek Soo Kim, Moon-Woo Seong, Jae Hyeon Park
Ann Clin Microbiol 2026 March, 29(1):2. Published on 27 February 2026.
Kibum Jeon, Nuri Lee, Hyun Soo Kim, Han-Sung Kim, Wonkeun Song, Jae-Seok Kim
Ann Clin Microbiol 2026 March, 29(1):1. Published on 6 March 2026.
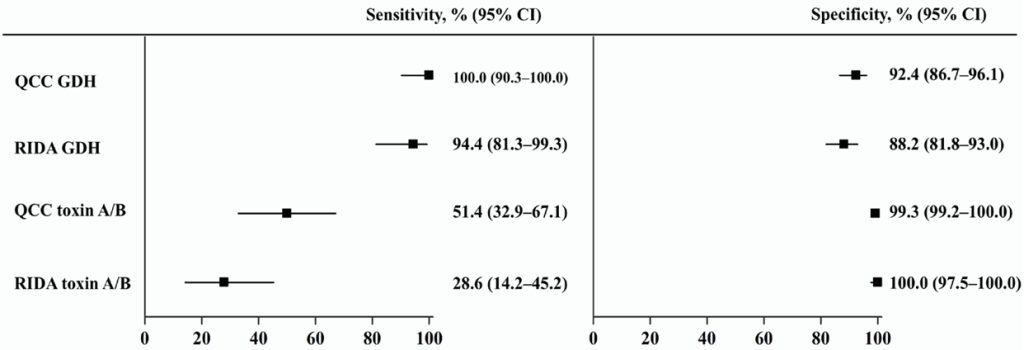
Background: Enzyme immunoassays (EIAs), which detect glutamate dehydrogenase (GDH) and toxin A/B, are widely used to screen for Clostridioides difficile infection (CDI); however, their sensitivity is lower than that of molecular assays. This study aimed to evaluate the performance of two EIAs, C. Diff Quik Chek Complete (QCC) and RIDASCREEN (RIDA), and investigate the cycle threshold (Ct) values from two real-time polymerase chain reaction (PCR) assays (Allplex GI–Bacteria(I) and Xpert C. difficile) in EIA-discordant samples.
Methods: A total of 180 clinical stool samples were tested using QCC, RIDA, and Allplex GI-Bacteria(I) PCR assays. The Xpert C. difficile assay was used to analyze discordant results.
Results: QCC and RIDA showed high sensitivities for GDH detection, 100.0% and 94.4%, respectively. QCC was significantly more sensitive than RIDA for toxin detection (51.4% vs. 28.6%, p = 0.007). In 25 EIA-discordant, Xpert positive samples, the Ct values of the toxin B gene ranged from 31.5 to 44.8 (mean, 38.1) for Allplex PCR and from 23.7 to 36.3 (mean, 30.4) for Xpert PCR. The Ct values of the two PCR assays were not significantly correlated (r = 0.201, p = 0.324).
Conclusion: QCC is a suitable initial immunological test for diagnosing CDI. The lack of correlation in the Ct values between the two real-time PCR assays suggests that assay-specific validation is necessary for cutoff level interpretation.
Jungmi Kim, Inyoung Kang, Sunjoo Kim
Ann Clin Microbiol 2026 March, 29(1):3. Published on 9 March 2026.
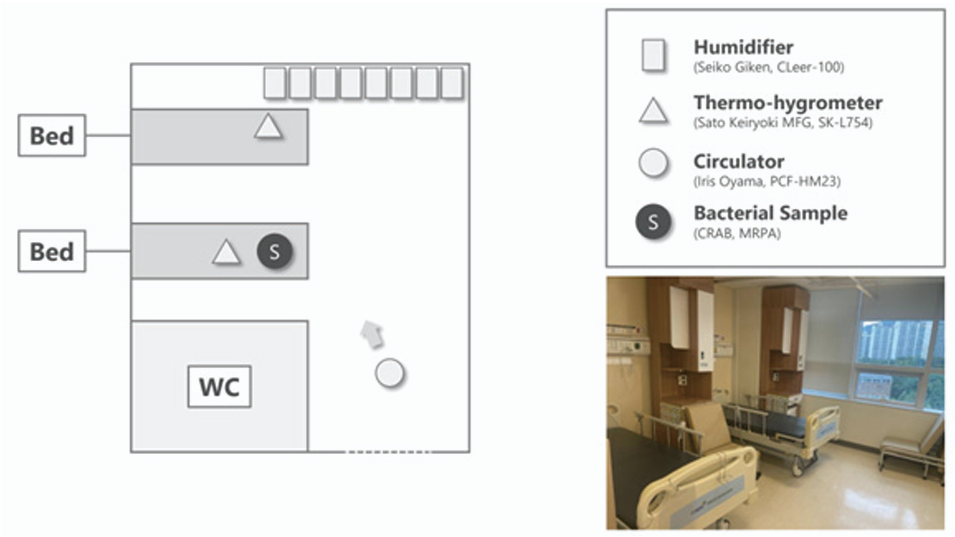
Background: Environmental contamination with multidrug-resistant organisms (MDROs), including carbapenem-resistant Acinetobacter baumannii (CRAB) and multidrug-resistant Pseudomonas aeruginosa (MRPA), remains a major challenge in healthcare facilities. Hypochlorous acid water (HOCl) has emerged as a promising disinfectant owing to its strong antimicrobial activity and favorable safety profile. This study aimed to evaluate the bactericidal efficacy of atomized HOCl against CRAB and MRPA in a hospital room.
Methods: An atomization experiment was conducted in a two-bed room. CRAB and MRPA were prepared using drying and non-drying methods, respectively. HOCl (CLFine) at concentration of 40 and 300 ppm was atomized using ultrasonic humidifiers. Bacterial samples were collected at 0, 1, 3, and 5 h after atomization. Viable bacterial counts were determined by culture, and bactericidal efficacy was evaluated.
Results: Atomized HOCl exhibited time- and concentration-dependent bactericidal effects against CRAB and MRPA. CRAB and MRPA reached their limits of detection at 3 and 5 h post-atomization at 40 ppm, and at 1 and 3 h at 300 ppm, respectively.
Conclusion: Atomized HOCl effectively inactivated CRAB and MRPA in a hospital room within 3–5 h. These findings support the potential application of HOCl atomization as an adjunctive environmental disinfection strategy for controlling MDRO contamination in healthcare facilities.
Special Issue on Current Trends in Laboratory Diagnosis and Public Health of Mycobacterial Diseases in Korea.
Hae-Sun Chung
Ann Clin Microbiol 2025 December, 28(4):27. Published on 20 December 2025.
Takashi Takahashi
Ann Clin Microbiol 2025 December, 28(4):22. Published on 1 December 2025.
In the present review, we systematically examine the diverse applications of whole-genome sequencing (WGS) and next-generation sequencing (NGS) to elucidate the evolution of clinical microbiology. The review aims to provide novel insight and to improve understanding of the applications of WGS in clinical microbiology laboratories. It is organized into the following sections: (1) the various types of NGS machines; (2) NGS workflows for obtaining genome sequences; (3) comparative genomic analysis; (4) RNA-seq (transcriptome) analysis; (5) genome-based bacterial typing; (6) genome-based antimicrobial resistance (AMR) detection; and (7) identification of integrative and conjugative elements carrying AMR gene(s). Four figures and three tables are provided to illustrate this information. The discussion focuses on WGS applications using several genera of microorganisms (Streptococcus, Enterococcus, Staphylococcus, Pasteurella, and Mycobacterium). Overall, WGS and related NGS technologies provide innovative clinical microbiology laboratory studies based on high-throughput genomic results for pathogen identification, tracking, and AMR/virulence profiling. In line with the concept of “One Health,” human and animal microbiology laboratories should pay careful attention to the dynamic evolution of WGS and related NGS technologies.
Chang-Ki Kim
Ann Clin Microbiol 2025 December, 28(4):26. Published on 20 December 2025.
(This article belongs to the Special Issue on Current Trends in Laboratory Diagnosis and Public Health of Mycobacterial Diseases in Korea.)
Tuberculosis (TB) remains a major global health threat, and the emergence and spread of drug-resistant Mycobacterium tuberculosis continue to undermine control efforts. Multidrug-resistant and rifampicin-resistant TB (MDR/RR-TB) is associated with prolonged treatment, higher toxicity, increased costs, and poorer outcomes compared to susceptible TB, making rapid and accurate drug susceptibility testing (DST) essential for effective patient management and transmission prevention. This review summarizes the current methods for DST in TB, focusing on the principles, strengths, and limitations of phenotypic and molecular approaches. Phenotypic DST, including the proportion, absolute concentration, and resistance ratio methods, and automated liquid culture systems, remains the conventional reference standard; however, conventional methods are limited by long turnaround times and technical complexity for certain drugs (such as pyrazinamide). Molecular DST targets resistance-associated mutations in key genes and is represented by line probe assays and cartridge-based platforms such as the Xpert MTB/RIF, which provide rapid results but are restricted to predefined genetic loci and may exhibit discordance with phenotypic DST, particularly with regard to borderline resistance. Next-generation sequencing (NGS)-based assays, including whole-genome sequencing and targeted NGS panels, offer comprehensive resistance profiling with high diagnostic accuracy and are increasingly being incorporated into international guidelines. Finally, we discuss the clinical interpretation of discordant results between genotypic and phenotypic DST, impact of revised rifampicin critical concentrations, and integration of DST results into contemporary World Health Organization guidelines and Korean treatment recommendations for MDR/RR-TB. Informed, methodologically grounded use of DST is crucial for optimizing the diagnosis and management of drug-resistant TB.
Sangsoo Jung, Eunsang Suh, Jun-Ki Lee, Byung Woo Jhun, Tae Yeul Kim, Hee Jae Huh, Nam Yong Lee
Ann Clin Microbiol 2025 December, 28(4):21. Published on 1 December 2025.
(This article belongs to the Special Issue on Current Trends in Laboratory Diagnosis and Public Health of Mycobacterial Diseases in Korea.)
Background: The growing burden of nontuberculous mycobacteria (NTM) raises concerns regarding cross-reactivity in molecular tuberculosis (TB) diagnostics. In the current study, we evaluated whether high NTM loads affect Mycobacterium tuberculosis (MTB) detection or rifampin (RIF) resistance calls using Xpert MTB/RIF (Xpert) and Xpert MTB/RIF Ultra (Xpert Ultra).
Methods: In vitro spiking experiments were performed by mixing eight NTM species (1 × 106 colony-forming unit [CFU]/mL) with heat-inactivated MTB (RIF-susceptible H37Rv; RIF-resistant S450L) at 5 × 103 CFU/mL in pooled smear-negative sputum, before testing them in parallel using Xpert and Xpert Ultra. We also retrospectively analyzed 334 results from lower respiratory specimens, including 32 NTM-positive specimens tested using both assays, and assessed the presence of probe amplification in the NTM-confirmed specimens.
Results: In spiking experiments, both assays showed no NTM-related cross-reactivity: RIF-susceptible mixes were “RIF resistance not detected,” S450L mixes were correctly resistant with preserved mutant melt peaks, and MTB-specific cycle threshold and melting peak temperature profiles were unchanged by NTM. Of the 334 clinical specimens, NTM was isolated from 32. Xpert classified all 32 as MTB-negative, and Xpert Ultra classified 31 of 32 as MTB-negative and one as “very low” positive in a patient with prior TB, consistent with residual nonviable DNA. In NTM-positive, Xpert/Xpert Ultra-negative specimens, neither assay showed probe amplification.
Conclusion: High-burden NTM did not compromise MTB detection or RIF-resistance determination using Xpert or Xpert Ultra. The assays demonstrated robust analytical specificity in mixed MTB〝NTM contexts, supporting their use where NTM carriage is common.
Jeong Su Park, Kyu-Hwa Hur, Woo Jin Shin, Hyunji Kim, Dong Woo Shin, Kyoung Un Park
Ann Clin Microbiol 2025 December, 28(4):24. Published on 20 December 2025.
(This article belongs to the Special Issue on Current Trends in Laboratory Diagnosis and Public Health of Mycobacterial Diseases in Korea.)
Background: Mycobacterium avium complex (MAC) is a major cause of pulmonary nontuberculous mycobacterial disease; however, treatment outcomes remain suboptimal. Phenotypic drug susceptibility testing (DST) is conditionally recommended; however, conventional broth microdilution is labor-intensive. The Sensititre SLOMYCO® panel offers a standardized platform for DST of slowly growing mycobacteria.
Methods: Eighty-six clinical MAC isolates (48 M. avium and 38 M. intracellulare) from respiratory specimens were tested for 13 antimicrobials using the SLOMYCO panel and reference Clinical and Laboratory Standards Institute (CLSI) broth microdilution methods at the Korean Institute of Tuberculosis. Essential agreement (EA) was defined as minimum inhibitory concentrations within ± 1 dilution, and categorical agreement (CA) was based on CLSI 2018 breakpoints for clarithromycin, amikacin, moxifloxacin, and linezolid.
Results: The EA was high for amikacin (90%), moxifloxacin (92%), linezolid (92%), and ethambutol (98%). Moderate EA was observed for clarithromycin (79%), ciprofloxacin (67%), and doxycycline (63%), and low EA was observed for trimethoprim-sulfamethoxazole (34%). The CA values were 100%, 77.9%, 69.8%, and 47.7% for clarithromycin, amikacin, moxifloxacin, and linezolid, respectively. All isolates were clarithromycin-susceptible according to both methods, and no clarithromycin- or amikacin-resistant isolates were detected.
Conclusion: The SLOMYCO DST system demonstrated high agreement with the reference methods for clarithromycin and amikacin in the tested susceptible population. The variability in the results for moxifloxacin and linezolid highlights the need for refined breakpoints. The validation of resistant isolates is essential before the SLOMYCO system can be recommended for comprehensive clinical applications.
Young Ah Kim, Jae Kwang Lee, Hee Kyoung Choi
Ann Clin Microbiol 2025 December, 28(4):20. Published on 20 November 2025.
Background: Diagnostic tests are essential for accurate disease identification and monitoring treatment responses. This study aimed to assess the factors influencing the requests for microbial diagnostic tests in patients with various infections.
Methods: Using tailored data from the National Health Insurance Big Data, we examined the usage patterns of microbiological tests among patients with pulmonary tuberculosis (TB) and major bacterial infections between 2020 and 2022, excluding overlapping and complex infections (n = 8,268,992). The types of bacterial infections included pneumonia, sepsis, urinary tract infections, and soft tissue infections, for which bacterial culture and antimicrobial susceptibility tests were performed. For pulmonary TB cases, acid-fast bacillus smears, mycobacterial cultures, and molecular diagnostic tests were performed. Multivariate analysis was used to identify the factors influencing the prescription of microbiological diagnostic tests, considering variables such as age, sex, Charlson comorbidity index, underlying disease, type of medical institution, residential area, insurance quintile, and disability status.
Results: Requests for TB-related and bacterial infection-related tests varied according to multiple factors, including sex, age, insurance quintile, residential area, presence or absence of disability, disease severity, type of medical institution admission, chemotherapy, steroid use, comorbid conditions, and underlying diseases.
Conclusions: This study provides a rare analysis of large-scale national data on various factors affecting test prescriptions, which could provide useful data for improving policies for appropriate test prescriptions.
Eun-Young Kim, Yeon Chan Choi, Hyeon Jin Choi, Si Hyun Kim, Jihyun Cho, Seok Hoon Jeong, Dokyun Kim, Hyun Soo Kim, Soo Hyun Kim, Young Ah Kim, Young Ree Kim, Nam Hee Ryoo, Jong Hee Shin, Kyeong Seob Shin, Young Uh, Jeong Hwan Shin
Ann Clin Microbiol 2025 December, 28(4):23. Published on 20 November 2025.
Background: Haemophilus influenzae is the causative pathogen for various infectious diseases, such as respiratory infections, otitis media, sinusitis, and meningitis. This study aimed to investigate the prevalence and molecular characteristics of β-lactam resistance in non-typeable H. influenzae isolates in South Korea.
Methods: In total, 115 non-duplicated H. influenzae isolates were included in this study. Bacterial identification and serotyping were performed using matrix-assisted laser desorption ionization-time of flight mass spectrometry and polymerase chain reaction (PCR) of bexA, respectively. Antimicrobial susceptibility was tested using the broth microdilution method. The production of β-lactamase was determined using nitrocefin disks. The presence of blaTEM and blaROB was confirmed using PCR. ftsI was analyzed to identify amino acid mutations in penicillin-binding protein (PBP) 3.
Results: Resistance rates to ampicillin, amoxicillin–clavulanate, and cefuroxime were 67.8%, 13.9%, and 32.2%, respectively. None of the isolates were resistant to cefotaxime or ceftriaxone. Among 78 ampicillin-resistant isolates, 71 were β-lactamase-producing ampicillin-resistant (BLPAR), and 7 were β-lactamase-non-producing ampicillin-resistant. All BLPAR isolates carried blaTEM, and none carried blaROB. Among 16 amoxicillin–clavulanate-resistant isolates, 15 β-lactamase producers harbored blaTEM. Four to 7 PBP3 mutations per isolate were detected in all 16 non-β-lactamase-producing ampicillin-resistant or cephalosporin-resistant isolates.
Conclusion: β-lactam resistance in non-typeable H. influenzae isolates is highly prevalent in South Korea, primarily because of blaTEM and various PBP3 mutations. Therefore, continuous monitoring of antimicrobial resistance rates and mechanisms in non-typeable H. influenzae is necessary.
Sungwook Song, Eunsim Shin, Sunghee Han, Sang Gon Lee
Ann Clin Microbiol 2025 December, 28(4):25. Published on 20 November 2025.
Background: Human metapneumovirus (hMPV) is a major cause of acute respiratory infections in children and adults worldwide; however, no antiviral therapies or vaccines are currently available. Therefore, rapid and reliable diagnostic tools are required to support timely patient management and control outbreaks.
Methods: We retrospectively evaluated GenBody hMPV Ag Rapid Test results using 165 consecutive clinical samples collected from patients with suspected respiratory infections between May and August 2024. Specimens were pre-characterized using a confirmatory reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay. Diagnostic performance was summarized using percent positive agreement (PPA), percent negative agreement (NPA), and overall percent agreement (OPA), with subgroup analyses by age and cycle threshold (Ct) groups.
Results: The GenBody assay achieved a PPA of 92.7% (51/55), NPA of 100% (110/110), and OPA of 97.6% (161/165), with no false-positive results. Age-stratified analysis showed high PPA in infants (93.3%) and children (97%), whereas estimates in hMPV-positive adolescents (n = 2) and adults (n = 5) were less precise owing to the small sample size. In the Ct-stratified analysis, PPA was 100% for specimens with Ct ≤ 28 (39/39), whereas all false-negative results occurred in the Ct > 28 group (12/16).
Conclusion: In this retrospective, single-center evaluation, the GenBody hMPV Ag Rapid Test showed high agreement with RT-qPCR, with reduced detection in specimens with higher Ct values. Further prospective studies and long-term stability assessments are warranted to confirm the performance across a broader range of clinical settings and storage conditions.
For Authors
Publisher
Subscribers
for quarterly newsletter subscription & instant discount on publication fee!
Sponsors