Table 3. PBP3 mutation profiles of β-lactamase-non-producing β-lactam-resistant Haemophilus influenzae
Ann Clin Microbiol 2025;28(4):23. Prevalence and molecular characteristics of β-lactam resistance in non-typeable Haemophilus influenzae isolates in Korea Download table Resistance profile Isolates (n = 16) TEM ftsI allele Amino acid substitutions D350 S357 M377 S385 L389 I449 A502 R517 N526 AM 2 – 40 N N I T F K AM, […]
Fig. 4. Distribution of negative samples by age group.
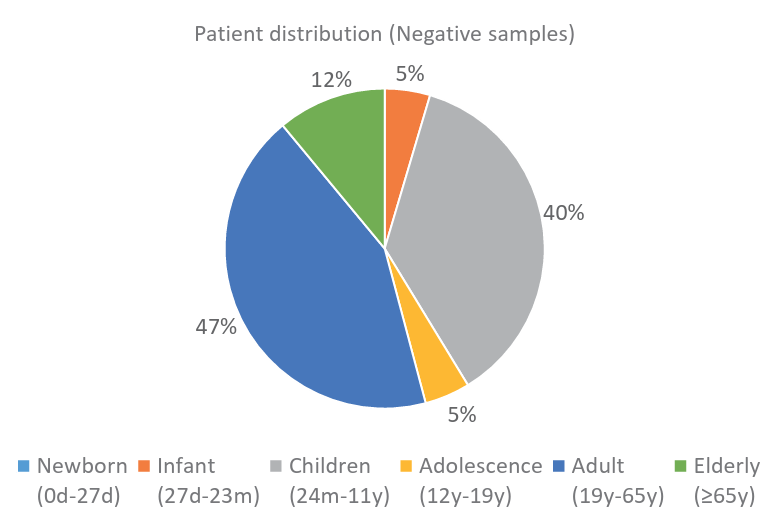
Ann Clin Microbiol 2025;28(4):25. Clinical evaluation of GenBody hMPV Ag Rapid Test: a diagnostic accuracy study Download image
Fig. 3. Distribution of positive samples by age group.
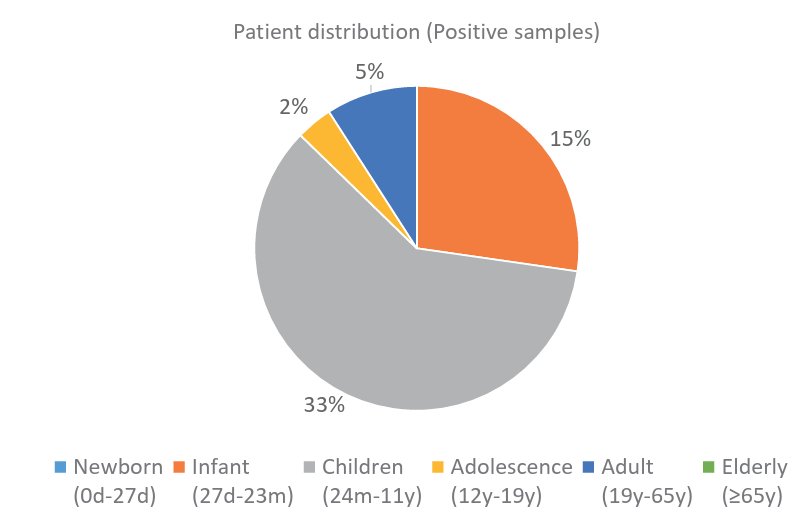
Ann Clin Microbiol 2025;28(4):25. Clinical evaluation of GenBody hMPV Ag Rapid Test: a diagnostic accuracy study Download image
Fig. 2. Distribution of sample count based on Ct value. Ct, cycle threshold.
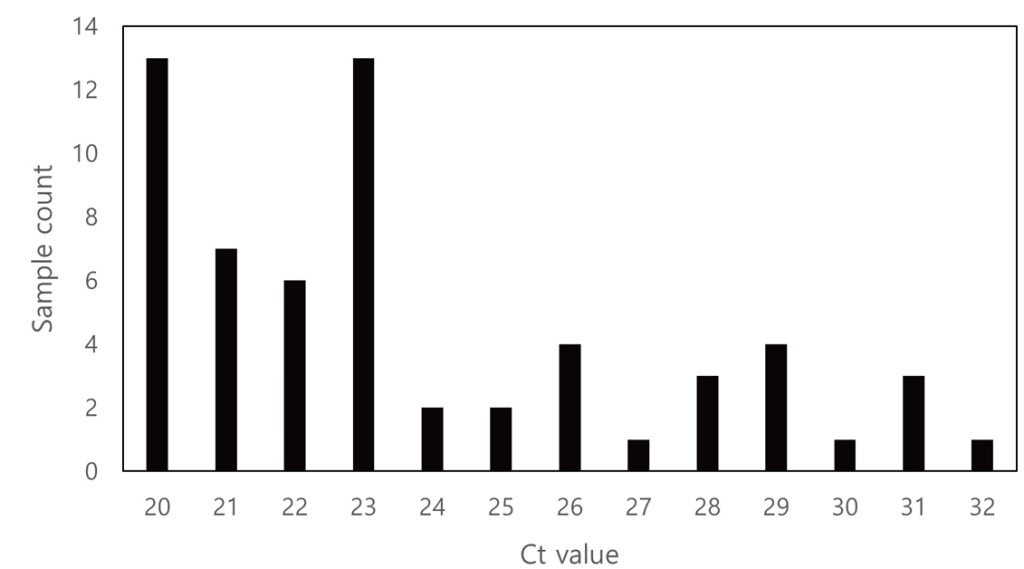
Ann Clin Microbiol 2025;28(4):25. Clinical evaluation of GenBody hMPV Ag Rapid Test: a diagnostic accuracy study Download image
Fig. 1. Outline of the testing process for performance evaluation. PCR, polymerase chain reaction; hMPV, human metapneumovirus; ID, identification; IRB, institutional review board; RT-qPCR, reverse transcription-quantitative PCR; PPA, percent positive agreement; NPA, percent negative agreement; OPA, overall percent agreement; Ct, cycle threshold.
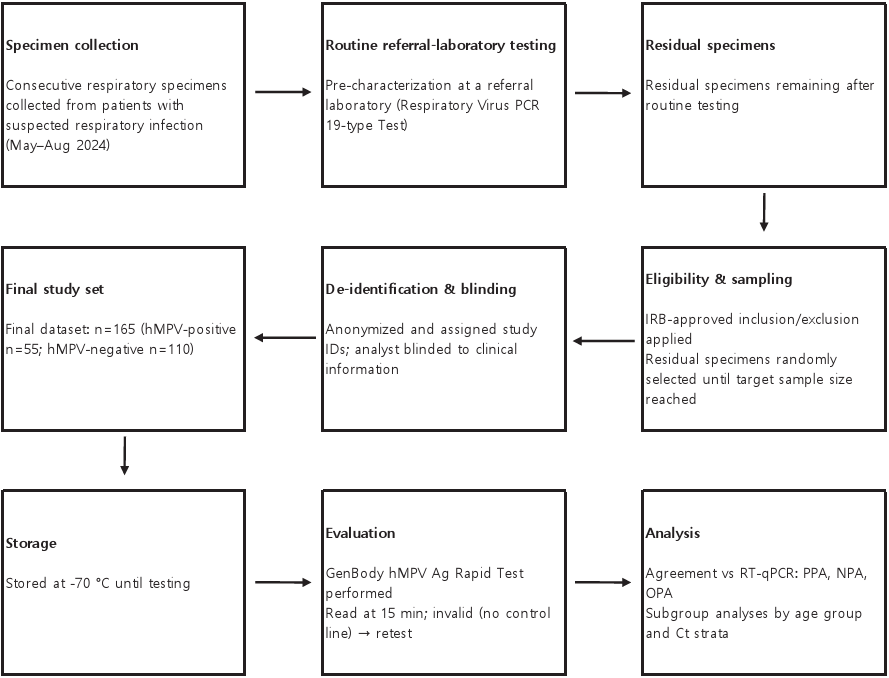
Ann Clin Microbiol 2025;28(4):25. Clinical evaluation of GenBody hMPV Ag Rapid Test: a diagnostic accuracy study Download image
Table 5. Sample distribution according to Ct values
Ann Clin Microbiol 2025;28(4):25. Clinical evaluation of GenBody hMPV Ag Rapid Test: a diagnostic accuracy study Download table Ct strata Ct values Ct < 24 24 ≤ Ct ≤ 28 28 < Ct Sample distribution 54.54% (30/55) 16.36% (9/55) 29.09% (16/55) Sensitivity 100% (30/30) 100% (9/9) 75% (12/16) Abbreviation: Ct, cycle threshold
Table 4. Positive sample distribution according to age
Ann Clin Microbiol 2025;28(4):25. Clinical evaluation of GenBody hMPV Ag Rapid Test: a diagnostic accuracy study Download table Age group Distribution (positive sample) Sensitivity Newborn (≤27 d) 0% (0/55) – Infant (28 d–23 m) 27.3% (15/55) 93.3% (14/15) Children (24 m–11 y) 60.0% (33/55) 97.0% (32/33) Adolescence (12 y–18 y) 3.6% (2/55) 50.0% (1/2) Adult […]
Table 3. Overall performance of the GenBody hMPV Ag Rapid Test
Ann Clin Microbiol 2025;28(4):25. Clinical evaluation of GenBody hMPV Ag Rapid Test: a diagnostic accuracy study Download table Variables Confirmatory method Total Positive Negative Positive 51 0 51 Negative 4 110 114 Total 55 110 165 Overall percent agreement (OPA) 97.58% (95% CI: 93.93%–99.05%) Percent positive agreement (PPA) 92.73% (95% CI: 82.74%–97.14%) Percent negative agreement […]
Table 2. Age distribution categories
Ann Clin Microbiol 2025;28(4):25. Clinical evaluation of GenBody hMPV Ag Rapid Test: a diagnostic accuracy study Download table Group Age Sample Count Percentage (%) Newborn Birth to 27 days 0 0 Infant 28 days to 23 months 19 11.5 Children 24 months to 11 years 75 45.5 Adolescence 12 years to 18 years 7 4.2 […]
Table 1. List of pathogens tested for cross-reactivity
Ann Clin Microbiol 2025;28(4):25. Clinical evaluation of GenBody hMPV Ag Rapid Test: a diagnostic accuracy study Download table No. Pathogen name No. Pathogen name No. Pathogen name 1 Adenovirus type 3 11 Parainfluenza virus 4a 21 Streptococcus pneumoniae 2 Adenovirus type 4 12 Parainfluenza virus type 1 22 Streptococcus pyogenes 3 Adenovirus type 7 13 […]
