1Smart Farm Research Center, Korea Institute of Science and Technology, Gangneung, Korea 2Department of Food Science and Biotechnology, Seoul National University of Science and Technology, Seoul, Korea
Corresponding to Eun Ha Lee, E-mail: ehlee@kist.re.kr
Ann Clin Microbiol 2025;28(1):2. https://doi.org/10.5145/ACM.2025.28.1.2
Received on 10 October 2024, Revised on 21 November 2024, Accepted on 27 November 2024, Published on 30 December 2024.
Copyright © Korean Society of Clinical Microbiology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND) (https://creativecommons.org/licenses/by-nc-nd/4.0/).
Purpose: The human gut mycobiome comprises diverse fungal species and plays a crucial role in health and disease, despite its relatively low abundance compared to bacterial populations. This review provides an overview of the mycobiome’s composition, developmental patterns, and dysbiosis in various pathological conditions. In addition, the complex interactions of fungal communities within the gut microbiome are discussed.
Current content: The development of the gut mycobiome follows patterns similar to those of the bacterial microbiome, with birth mode, diet, and age being key determinants. In contrast to the bacterial trends, mycobiome diversity increases during childhood and old age. Recent studies have revealed variations in the mycobiome composition across different ethnic groups. Mycobiome dysbiosis is associated with autoimmune, gastrointestinal, and cardiovascular diseases. Certain fungi, notably Candida albicans, are relatively more abundant in pathological states. Fungal metabolic activity, particularly secondary metabolite production, can significantly affect disease progression. Bacterial–fungal interactions in the gut microbiome are complex and modulated by environmental factors, such as diet and antibiotic use. Moreover, the gut mycobiome modulates therapeutic efficacy. Gut fungi enhance the bioactivity of compounds derived from natural products through biotransformation, including their anticancer and anti-inflammatory effects. This suggests the potential of the gut mycobiome to optimize the therapeutic efficacy of natural products.
Conclusion: This review highlights the relevance of the gut mycobiome as both a diagnostic biomarker and a therapeutic target. Future research should focus on elucidating the causal relationships between mycobiome alterations and disease states, and further explore bacterial–fungal interactions within the gut ecosystem.
Biotransformation, Candida albicans, Dysbiosis, Microbial interactions, Mycobiome
Humans coexist with a diverse array of microorganisms. These microorganisms have significant roles in the environment, food, and the human body. Human microbiota comprises various microbial species, including bacteria, archaea, and fungi, which can interact with the host in ways that may contribute to health maintenance or disease pathogenesis [1,2].
The gut microbiota profoundly affects human metabolism, immune responses, and overall health [3,4]. Gut bacteria are crucial for synthesizing vitamins, metabolizing nutrients, and regulating immune functions [5]. These findings suggest that gut bacteria are essential for maintaining human health and have significant potential as biological markers for diagnosing and treating specific diseases. The predominant focus of research on the gut microbiome has been bacteria. Gut fungi have received relatively little attention. Analysis of fecal samples to characterize the gut microbiome revealed a substantially lower fungal cell count (approximately 105 cells/g feces) than bacterial cell count (approximately 1012 cells/g feces) [6].
Recent advancements in next-generation sequencing (NGS) technologies have spurred increased interest in mycobiome research by providing methods for the rapid and accurate analysis of diverse microbial species. NGS has become indispensable for elucidating the mechanisms and interactions of fungal communities and their association with diseases [7].
The understanding of the interactions between mycobiome and disease pathogenesis remains limited. Elucidating the effect of fungi on disease onset and diagnostic potential remains challenging. Multifaceted approaches, including the integration of metagenomic, transcriptomic, and metabolomic analyses, are required for a more comprehensive understanding.
This review systematically explores the diverse roles of the human mycobiome in health, addresses current research limitations, and proposes future directions. The information is expected to provide novel insights for the development of diagnostic and therapeutic strategies based on mycobiome research in clinical settings.
The human mycobiome is comprised of diverse fungal species that inhabit various body sites, including the skin, gut, oral cavity, and respiratory tract (Fig. 1) [8]. Fungal genes constitute only 0.1% of the total microbial genes in the gut, as shown in early shotgun metagenomic sequencing studies [9]. Nonetheless, the impact of fungal communities on human health is substantial because of their unique physiological and metabolic properties [10]. Despite the relatively low abundance of fungi compared to bacteria, these microorganisms may play critical roles in maintaining host health and homeostasis, mainly because of their large size and different metabolic functions [11].
Distinct fungal communities reside in different anatomical sites and contribute to local and systemic health. Notably, the gut harbors the highest density of fungi compared to other body regions. Early human mycobiome studies identified Saccharomyces, Candida, and Cladosporium as the dominant fungal genera in fecal samples [10]. However, there are considerable interethnic variations in gut mycobiome composition. For instance, analysis of fecal samples from 135 healthy Chinese individuals identified 12,453 fungal isolates, predominantly from the genera Aspergillus, Penicillium, Talaromyces, Candida, and Pichia. This distinct distribution contrasts with findings from Western populations [12].
The mode of delivery significantly influences the initial colonization of the neonatal gut mycobiome. Newborns delivered via vaginal birth tend to have mycobiome profiles that resemble those of their mother’s vaginal mycobiome. In contrast, those born via cesarean section acquire fungal communities mainly from environmental sources and maternal skin. The mycobiome is shaped by close contact with caregivers, including mothers, during early infancy [13–16].
The composition of the gut mycobiome undergoes dynamic changes at different life stages. In early infancy, the mycobiome is dominated by Saccharomycetales and Malasseziales species. As dietary patterns shift with the introduction of solid foods, species such as Saccharomyces cerevisiae, Cystofilobasidium spp., Ascomycota spp., and Monographella spp. become more prevalent [17]. Alterations in fungal communities have been strongly correlated with dietary diversification and nutritional complexity. Western diets are characterized by a high consumption of bread, beer, and dairy products. Saccharomyces tends to dominate, whereas Aspergillus is frequently detected in vegetarians [18,19]. Growth of Candida is promoted by diets enriched in carbohydrates and decreased by diets rich in amino acids, fatty acids, and proteins [20].
Interestingly, the diversity of the gut mycobiome is modulated by age, with increased diversity during childhood and old age. This pattern contrasts with the trends observed in the bacterial microbiome. While bacterial diversity typically decreases with aging, fungal diversity increases, potentially impacting immune responses and inflammation [10,21].
Gut mycobiome dysbiosis has been implicated in various pathological conditions including autoimmune disorders, gastrointestinal diseases, respiratory infections, dermatological conditions, and even cancer [17]. Disruption of fungal communities perturbs homeostasis of the gut microbiota, altering host immune responses and increasing susceptibility to numerous diseases. Early studies described disease-associated gut mycobiome dysbiosis by examining the ratio between the phyla Basidiomycota and Ascomycota. These differences suggest that specific fungal phyla play crucial roles in disease development and progression. Notably, patients with colorectal cancer and inflammatory bowel disease demonstrate a statistically significant increase in the ratio of Basidiomycota to Ascomycota compared to healthy controls, reflecting dynamic alterations in fungal community composition during disease states [22,23].
Candida albicans is a common fungal species in the gut. Increased relative abundance of C. albicans in pathological conditions, including liver cirrhosis, ulcerative colitis, and coronavirus disease 2019, has been demonstrated [11,17,23,44–55] (Table 1). C. albicans secretes gliotoxins, which are potent secondary metabolites that disrupt the immune defense and promote disease progression [24]. The dysregulated production of these metabolites may induce local inflammation and alter the microbial community structure, exacerbating disease severity. Analysis of metadata from patients with autoimmune diseases revealed that many patients had utilized immunosuppressants to manage symptoms. Notably, those treated with diseasemodifying antirheumatic drugs and inhibitors of tumor necrosis factor-alpha exhibited an impaired C. albicans-specific Th17 immune response, which is believed to contribute to the increased prevalence of C. albicans [25]. Therefore, knowledge of the pathogenic mechanisms and changes in gut fungal communities is essential for understanding the role of mycobiomes in various diseases and identifying novel therapeutic targets.
A comprehensive analysis of metabolite profiles from fecal samples of patients with specific diseases revealed that the pathological effects of gut fungi are primarily mediated by secondary metabolites [12]. These metabolites modulate both local and systemic immune responses, alter metabolic pathways, and influence gut-brain communication through the neuro-immune–endocrine axis. Consequently, patients with ulcerative colitis have a 10% increased risk of developing Alzheimer’s disease, whereas those with Crohn’s disease have a 2.7-fold higher risk of developing frontotemporal dementia [24]. Saccharomyces cerevisiae is typically abundant in healthy human guts and even more abundant in children with autism spectrum disorder [26]. These findings suggest that gut mycobiome alterations may vary depending on the disease context, and that specific fungal species may exhibit divergent behaviors under different pathological conditions.
The foregoing findings highlight that understanding dynamic shifts in fungal community composition is crucial for recognizing the potential of the mycobiome as a diagnostic biomarker and therapeutic target. Observations, such as those in children with autism spectrum disorder, underscore the necessity of considering disease-specific mycobiome shifts and their potential role in modulating health and disease outcomes. Future studies should focus on examining drug-induced alterations in fungal communities to better understand disease pathogenesis.
Table 1. Disease-specific alterations in the gut mycobiome
| Diseases | Increase | Decrease | References |
|---|---|---|---|
| Crohn’s disease | Aspergillus clavatus, Candida albicans, C. tropicalis, C. glabrata, C. lusitaniae, Cryptococcus neoformans, Cyberlindnera jadinii, Debaryomyces hansenii, Kluyveromyces marxianus | Saccharomyces cerevisiae | [17, 44] |
| Ulcerative colitis | C. albicans, Debaryomyces | Aspergillus flavus, A. cibarius, C. soja, D. hansenii | [17] |
| Irritable bowel syndrome | Cladosporium, Malassezia, C. albicans, S. cerevisiae | Aspergillus, Mycosphaerella, Sporidiobolus, Pandora | [17] |
| Intestinal GVHD | C. albicans, C. parapsilosis | [17] | |
| Rheumatoid arthritis | C. albicans | Aspergillus | [45] |
| Ankylosing spondylitis | Alternaria, Candida, Chytridiomycota, Glomeromycota | Amphinema, Clavulina | [46] |
| Colorectal cancer | Malassezia mycetomatis, Malassezia, Trichosporon, A. ramulosa, C. tropicalis | Saccharomyces | [11, 17, 23, 47, 48] |
| Pancreatic cancer | Malassezia | [17] | |
| Gastric carcinogenesis | Alternaria, Candida | Saitozyma, Thermomyces | [17] |
| Alcoholic liver disease | Candida | Debaryomyces, Epicoccum, Galactomyces, Penicillium | [17] |
| Liver cirrhosis | Candida | [17] | |
| Primary sclerosing cholangitis | Candida, Exophiala, Humicola | Saccharomyces cerevisiae | [17] |
| Clostridioides difficile infection | Aspergillus, Cladosporium, C. albicans | [17] | |
| COVID-19 infection | C. albicans, C. auris, C. glabrata | A. flavus | [17] |
| Hepatitis B virus infection | Aspergillus, Candida, Galactomyces, Saccharomyces | Chaetomium | [49] |
| HIV infection | Candida, C. albicans | [50, 51] | |
| Obesity | Candida, Nakaseomyces, Penicillium, Pichia | Mucor, Wallemia, Bettsia | [52] |
| Type 1 diabetes | Candida, Saccharomyces | [17] | |
| Type 2 diabetes | Aspergillus, Candida, Cladosporium, Kodamaea, Meyerozyma, Mortierella | Mucor, Mucor restricta | [17] |
| Atherosclerosis | Mucor racemosus, Mucor restricta | [17] | |
| Multiple sclerosis | Aspergillus, Saccharomyces | [53] | |
| Rett syndrome | Candida | A. versicolor | [17] |
| Autism spectrum disorder | S. cerevisiae | [17] | |
| Schizophrenia | Chaetomium | Trichoderma | [54, 55] |
| Alzheimer’s disease | C. albicans, S. cerevisiae, Botrytis, Cladosporium, Kazachstania, Phaeococcomyces, C. tropicalis, Schizophyllum commune | Meyerozyma, Rhodotorula mucilaginosa | [17] |
Abbreviations: GVHD, graft-versus-host disease; COVID-19, coronavirus disease 2019.
Fungi are significant opportunistic pathogens in immunocompromised individuals and integral components of the gut microbiome. These microorganisms are crucial in maintaining gut homeostasis. Previous studies have revealed a competitive relationship between gut bacterial and fungal populations, where fungal overgrowth commonly occurs following the disruption of bacterial equilibrium (Fig. 2). This interaction is modulated by direct bacterial antagonism or regulation of host immune responses through bacterial metabolites [10].
Bacterial–fungal interactions may manifest as competition for nutrients and colonization sites, or as cooperative behaviors that promote mutual growth, depending on the type of disease and environmental factors. For example, there is a stronger correlation between fungal and bacterial populations in patients with ulcerative colitis, but not in those with Crohn’s disease [22]. Similarly, a high-fat diet significantly reduces bacterial–fungal correlations in the gut microbiome of mice [27].
Bacterial–fungal interactions can also influence therapeutic outcomes. For instance, the presence of C. albicans reduces the efficacy of fecal microbiota transplantation in treating Clostridioides difficile infections. In a dextran sulfate sodium-induced colitis model, the removal of Enterobacteriaceae mitigated the pathogenic effects of C. albicans and nullified the beneficial effects of Saccharomyces boulardii [17].
Iron acquisition is another critical factor in bacterial–fungal interactions. C. albicans and Cryptococcus neoformans are unable to synthesize their own siderophores and rely on siderophores produced by neighboring bacteria. This phenomenon, known as “iron parasitism,” enhances fungal growth and pathogenicity. Conversely, the bacterial production of weak organic acids lowers the environmental pH and suppresses fungal growth. Short-chain fatty acids, such as butyrate, reportedly inhibit the growth of certain pathogenic fungi and attenuate their virulence [28]. An illustrative example of bacterial metabolites influencing fungal growth is the production of 1-acetyl-beta-carboline by Lactobacillus species, which inhibits the Dual-specificity tyrosine-phosphorylation-regulated kinase 1 family member Yak1. The inhibition of this kinase impedes biofilm formation by C. albicans and interferes with the yeast-to-filamentous transition, thereby reducing the pathogenic potential of C. albicans [29].
Bacterial–fungal interactions are also regulated through quorum sensing systems. Supernatants from Lactiplantibacillus plantarum cultures reportedly inhibited the growth of S. cerevisiae. Additionally, coculturing L. plantarum with S. cerevisiae suppressed the growth of S. cerevisiae. Transcriptomic analysis of L. plantarum and S. cerevisiae revealed that the lamBDCA quorum sensing system in L. plantarum was activated, whereas physiological and growth-related pathways in S. cerevisiae were downregulated. These results suggest that L. plantarum uses extracellular metabolites to mediate interspecies signaling, thereby inhibiting the growth of S. cerevisiae [30].
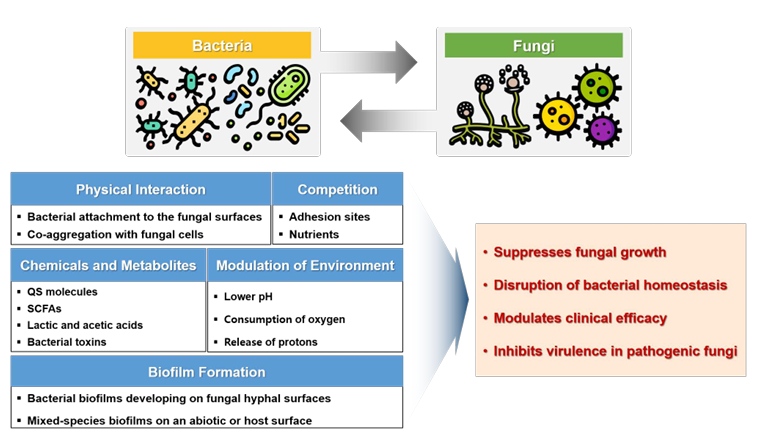
Fig. 2. Mechanisms and consequences of bacterial–fungal interactions. Bacterial and fungal species modulate fungal growth in human hosts via diverse mechanisms. QS, quorum sensing; SCFA, shortchain fatty acid.
The gut mycobiome composed of diverse fungal species is critical for altering the chemical structure and bioactivity of various drugs and natural compounds through biotransformation processes. Fungi use diverse enzymatic systems to metabolize xenobiotics, natural products, and pharmaceutical agents, thereby influencing drug efficacy and safety.
Fungal CYP enzymes exhibit broad substrate specificity, which allows the conversion of complex chemical structures to metabolites with modified pharmacological properties. For example, Pleurotus ostreatus possesses CYP monooxygenases that catalyze hydroxylation reactions on aromatic substrates, which are crucial for the detoxification and modification of drugs such as carbamazepine [31]. These CYPmediated transformations are similar to those observed in mammalian liver cells, and produce metabolites with analogous biological effects [32].
The gut mycobiome has the remarkable ability to biotransform natural products and generate metabolites with enhanced biological activity. Pichia anomala isolated from human fecal samples is capable of converting curcumin to hexahydrocurcumin and octahydrocurcumin, which exhibit enhanced antitumor activity compared to the parent compound [24,33,34]. Ginsenoside Rg1 is converted to 20(S)-protopanaxatriol by gut bacteria. The resulting molecule undergoes further metabolism by the Mucor spinosus AS 3.3450 fungus, generating new metabolites that exhibit superior anticancer activity compared with the parent compound 20(S)-protopanaxatriol [35]. The bioactive compound dihydrocapsaicin derived from Capsicum spp. is biotransformed by gut fungi (Aspergillus fumigatus, A. japonicus, and C. parapsilosis) in vitro, resulting in the production of metabolites with potent inhibitory effects on lysine-specific demethylase 1 [36]. Such biotransformation processes underscore the role of fungi in modulating the bioactivity of natural compounds, thereby influencing their therapeutic outcomes.
A. niger metabolizes 18β-glycyrrhetinic acid into a novel metabolite with increased anti-inflammatory effects [37]. Fungus-mediated transformation of chenodeoxycholic acid by Rhizopus microsporus generates metabolites that act as antagonists of the farnesoid X receptor. This receptor is crucial in bile acid metabolism and glucose homeostasis [38]. The findings suggest that gut fungi have the potential to affect host metabolic health and contribute to the onset of metabolic disorders. The ability of gut fungi to perform regio- and stereoselective transformations makes them ideal biocatalysts for the synthesis of pharmaceutical intermediates. S. cerevisiae has been reported to perform the regioselective reduction of complex substrates, such as the conversion of tert-butyl 6-chloro-3,5-dioxohexanoate to hydroxy keto esters, which are key intermediates in statin synthesis. These unique metabolic processes are essential for optimizing drug synthesis and enhancing therapeutic properties [39].
The gut mycobiome can modify the chemical structures of drugs and natural compounds, affecting their pharmacokinetics as well as host health and disease progression. These biotransformation processes may induce drug–drug or drug–microbe interactions, potentially altering therapeutic efficacy. A comprehensive understanding of fungal biotransformation mechanisms and their roles in drug metabolism is required. Future research should focus on identifying specific fungal enzymes and metabolic pathways involved in these processes to leverage their potential for personalized medicine and innovative drug development.
The gut mycobiome includes a diverse range of fungal species. Methodological limitations associated with culture-dependent techniques have restricted studies of the gut mycobiome. These methods capture only a fraction of the fungal diversity, leading to an incomplete understanding of the role of fungi in the gut microbiome. NGS technologies have revolutionized the field by enabling high-throughput sequencing and comprehensive profiling of fungal communities [40,41]. However, several technical and analytical challenges must be addressed in mycobiome research.
One of the primary limitations is the low abundance of fungal sequences in the gut microbiome relative to bacterial DNA. This underrepresentation makes it difficult to accurately identify fungal species and determine their functional roles. Moreover, contamination from host DNA complicates data interpretation, leading to potential biases in the assessment of fungal diversity and ecological relevance [11]. The incomplete nature of the current fungal genome reference databases exacerbates these issues, as many fungi still need to be characterized at the genomic level. Such deficiencies hinder precise species-level classification, functional prediction, and studies of interspecies interactions and metabolic pathways [42]. Current NGS methods such as amplicon sequencing and whole-metagenome shotgun sequencing have significantly advanced our understanding of the mycobiome. However, they still need to detect low abundance fungi because of their reliance on reference genomes and the challenges in assembling fragmented sequences. Additionally, the high cost and computational complexity of NGS methods restrict large-scale studies, thereby reducing statistical power and reproducibility. To address these limitations, the development of more cost-effective sequencing platforms, such as Oxford Nanopore and Illumina MiSeq, along with innovations in bioinformatics tools, is critical [43]. Future research should aim to refine sequencing technologies to increase their sensitivity and accuracy. Techniques such as digital polymerase chain reaction and deep whole-metagenome sequencing offer potential solutions for detecting low abundance fungal species. Moreover, combining NGS with other omics approaches that include metatranscriptomics, metaproteomics, and metabolomics will enable a more comprehensive understanding of fungal functions within the gut ecosystem. Integrative approaches are essential for elucidating the metabolic interactions between mycobiomes, bacteriomes, and host cells to provide insights into how fungal metabolites influence health and disease outcomes. Another critical area of focus is the expansion and improvement of fungal genome reference databases. Comprehensive databases will facilitate more accurate taxonomic classification and functional annotation, supporting a deeper understanding of fungal diversity and metabolic capabilities [17,42]. The goal of expanding and improving fungal genome reference databases can be facilitated through the de novo genome assembly of uncharacterized fungal species, along with efforts to catalog fungal diversity across various environments. Addressing these challenges requires interdisciplinary collaboration and the adoption of advanced bioinformatics tools. Developing standardized workflows for mycobiome data analysis, integrating machinelearning-based algorithms, and creating publicly accessible databases will enhance the reproducibility and reliability of mycobiome research. Furthermore, integrating NGS with metabolomics and transcriptomics will advance our knowledge of how gut mycobiomes affect host health. Linking fungal gene expression profiles with metabolic output may provide novel insights into the role of fungi in modulating host metabolism and immune responses. Evidence from these studies indicates that specific fungal metabolites such as those produced through bile acid biotransformation play critical roles in the pathogenesis of metabolic disorders, including obesity and diabetes [38]. The final step in integrating mycobiome research into clinical practice is the development of diagnostic tools and therapeutic strategies based on fungal biomarkers. Specific fungal taxa or metabolites can serve as disease markers, facilitating early diagnosis and personalized treatment. Furthermore, targeted therapies such as probiotics and antifungal agents have the potential to modulate disease progression and improve patient outcomes by manipulating the mycobiome.
Despite the low abundance of fungal species, the human gut mycobiome plays critical roles in human health and disease. The intricate interactions between fungal and bacterial communities, production of bioactive metabolites, and their influence on therapeutic efficacy have a significant impact on human health. To achieve a deeper understanding of the gut mycobiome, future research must address existing methodological limitations, expand fungal genome reference databases, and incorporate multiomics approaches. Advancements in these areas will pave the way for the identification of novel diagnostic biomarkers and therapeutic strategies to enhance disease management and promote overall health.
This was not a human population study. Therefore, institutional review board approval and informed consent were not required.
No potential conflicts of interest relevant to this article were reported.
None.
This review article does not involve the generation or analysis of new datasets. All data supporting the f indings are derived from previously published studies, which are appropriately cited within the manuscript.
1. Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct Target Ther 2022;7:1-28.


2. Matijašić M, Meštrović T, Čipčić Paljetak H, Perić M, Barešić A, Verbanac D. Gut microbiota beyond bacteria-mycobiome, virome, archaeome, and eukaryotic parasites in IBD. Int J Mol Sci 2020;21:2668.


3. Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013;14:685-90.


4. Wang H, Wei CX, Min L, Zhu LY. Good or bad: gut bacteria in human health and diseases. Biotechnol Biotechnol Equip 2018;32:1075-80.
5. LeBlanc JG, Milani C, De Giori GS, Sesma F, Van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol 2013;24:160-8.

6. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016;14:e1002533.


7. Tiew PY, Mac Aogain M, Ali NAtBM, Thng KX, Goh K, Lau KJ, et al. The mycobiome in health and disease: emerging concepts, methodologies and challenges. Mycopathologia 2020;185:207-31.


8. Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med 2013;5:1-12.


9. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59-65.


10. Kapitan M, Niemiec MJ, et al. eds. Fungi as part of the microbiota and interactions with intestinal bacteria. In: Rodrigues ML. Fungal physiology and immunopathogenesis. Switzerland; Springer, 2019:265-301.

11. Richard ML and Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 2019;16:331-45.

12. Yan Q, Li S, Yan Q, Huo X, Wang C, Wang X, et al. A genomic compendium of cultivated human gut fungi characterizes the gut mycobiome and its relevance to common diseases. Cell 2024;187:2969-89.

13. Bliss JM, Basavegowda KP, Watson WJ, Sheikh AU, Ryan RM. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr Infect Dis J 2008;27:231-5.

14. Nagata R, Nagano H, Ogishima D, Nakamura Y, Hiruma M, Sugita T. Transmission of the major skin microbiota, Malassezia, from mother to neonate. Pediatr Int 2012;54:350-5.

15. Juyal D, Sharma M, Pal S, Rathaur VK, Sharma N. Emergence of non-albicans Candida species in neonatal candidemia. N Am J Med Sci 2013;5:541-5.

16. Ward TL, Dominguez-Bello MG, Heisel T, Al-Ghalith G, Knights D, Gale CA. Development of the human mycobiome over the first month of life and across body sites. MSystems 2018;3:e00140-17.


17. Zhang F, Aschenbrenner D, Yoo JY, Zuo T. The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe 2022;3:e969-83.

18. Suhr MJ, Banjara N, Hallen-Adams HE. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol 2016;62:209-15.

19. Hallen-Adams H and Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence 2017;8:352-8.


20. Otašević S, Momčilović S, Petrović M, Radulović O, Stojanović N, Arsić-Arsenijević V. The dietary modification and treatment of intestinal Candida overgrowth-a pilot study. J Mycol Med 2018;28:623-7.

21. Strati F, Di Paola M, Stefanini I, Albanese D, Rizzetto L, Lionetti P, et al. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front Microbiol 2016;7:1227.


22. Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut 2017;66:1039-48.


23. Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019;68:654-62.


24. Aminudin NI, Amran NA, Zainal Abidin ZA, Susanti D. Biotransformation of curcumin and structure-activity relationship (SAR) of its analogues: a systematic review. Biocatal Biotransformation 2023;41:1-14.
25. Bishu S, Su EW, Wilkerson ER, Reckley KA, Jones DM, McGeachy MJ, et al. Rheumatoid arthritis patients exhibit impaired Candida albicans-specific Th17 responses. Arthritis Res Ther 2014;16:1-9.


26. Zou R, Wang Y, Duan M, Guo M, Zhang Q, Zheng H. Dysbiosis of gut fungal microbiota in children with autism spectrum disorders. J Autism Dev Disord 2021;51:267-75.

27. Heisel T, Montassier E, Johnson A, Al-Ghalith G, Lin YW, Wei LN, et al. High-fat diet changes fungal microbiomes and interkingdom relationships in the murine gut. mSphere 2017;2:e00351-17.


28. Lapiere A and Richard ML. Bacterial-fungal metabolic interactions within the microbiota and their potential relevance in human health and disease: a short review. Gut Microbes 2022;14:2105610.


29. MacAlpine J, Daniel-Ivad M, Liu Z, Yano J, Revie NM, Todd RT, et al. A small molecule produced by Lactobacillus species blocks Candida albicans filamentation by inhibiting a DYRK1-family kinase. Nat Commun 2021;12:6151.


30. Liu J, Huang TY, Liu G, Ye Y, Soteyome T, Seneviratne G, et al. Microbial interaction between Lactiplantibacillus plantarum and Saccharomyces cerevisiae: transcriptome level mechanism of cell-cell antagonism. Microbiol Spectr 2022;10:e01433-22.


31. Golan-Rozen N, Chefetz B, Ben-Ari J, Geva J, Hadar Y. Transformation of the recalcitrant pharmaceutical compound carbamazepine by Pleurotus ostreatus: role of cytochrome P450 monooxygenase and manganese peroxidase. Environ Sci Technol 2011;45:6800-5.

32. Martínez-Ramírez JA, Walther G, Peters FT. Studies on drug metabolism by fungi colonizing decomposing human cadavers. Part II: biotransformation of five model drugs by fungi isolated from post-mortem material. Drug Test Anal 2015;7:265-79.

33. Huang Y, Cao S, Zhang Q, Zhang H, Fan Y, Qiu F, et al. Biological and pharmacological effects of hexahydrocurcumin, a metabolite of curcumin. Arch Biochem Biophys 2018;646:317.

34. Zhang Z, Luo D, Xie J, Lin G, Zhou J, Liu W, et al. Octahydrocurcumin, a final hydrogenated metabolite of curcumin, possesses superior anti-tumor activity through induction of cellular apoptosis. Food Funct 2018;9:2005-14.

35. Zhang J, Guo H, Tian Y, Liu P, Li N, Zhou J, et al. Biotransformation of 20 (S)protopanaxatriol by Mucor spinosus and the cytotoxic structure activity relationships of the transformed products. Phytochemistry 2007;68:2523-30.

36. He X, Zhang B, Cao P, Wang H, Wu S, Wang G, et al. Biotransformation of dihydrocapsaicin by human intestinal fungi and the inhibitory effects of metabolites against LSD1. Heliyon 2022;8:e12325.


37. Zhang M, Zhang J, Wang C, Yan JK, Yi J, Ning J, et al. Biotransformation of 18β-glycyrrhetinic acid by human intestinal fungus Aspergillus niger RG13B1 and the potential anti-inflammatory mechanism of its metabolites. J Agric Food Chem 2022;70:1510415.

38. Wei X, Yao C, He X, Li J, Wang Y, Wang C, et al. Biotransformation of chenodeoxycholic acid by human intestinal fungi and the agonistic effects on FXR. Phytochemistry 2024;224:114162.

39. Bowman MJ, Jordan DB, Vermillion KE, Braker JD, Moon J, Liu ZL. Stereochemistry of furfural reduction by a Saccharomyces cerevisiae aldehyde reductase that contributes to in situ furfural detoxification. Appl Environ Microbiol 2010;76:4926-32.


40. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. PNAS 2012;109:6241-6.

41. Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017;5:1-13.


42. Xie Z and Manichanh C. FunOMIC: pipeline with built-in fungal taxonomic and functional databases for human mycobiome profiling. Comput Struct Biotechnol J 2022;20:3685-94.


43. Langsiri N, Worasilchai N, Irinyi L, Jenjaroenpun P, Wongsurawat T, Luangsa-Ard JJ, et al. Targeted sequencing analysis pipeline for species identification of human pathogenic fungi using long-read nanopore sequencing. IMA fungus 2023;14:18.


44. Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015;18:489-500.


45. Lee EH, Kim H, Koh JH, Cha KH, Lee KK, Kim WU, et al. Dysbiotic but nonpathogenic shift in the fecal mycobiota of patients with rheumatoid arthritis. Gut Microbes 2022;14:2149020.


46. Wang H, Wu H, Li KD, Wang YY, Huang RG, Du YJ, et al. Intestinal fungi and systemic autoimmune diseases. Autoimmun Rev 2023;22:103234.

47. Lin Y, Lau HCH, Liu Y, Kang X, Wang Y, Ting NLN, et al. Altered mycobiota signatures and enriched pathogenic Aspergillus rambellii are associated with colorectal cancer based on multicohort fecal metagenomic analyses. Gastroenterology 2022;163:908-21.

48. Wang T, Fan C, Yao A, Xu X, Zheng G, You Y, et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity 2018;49:504-14.e4.


49. Chen Y, Chen Z, Guo R, Chen N, Lu H, Huang S, et al. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis 2011;70:492-8.

50. Mukherjee PK, Chandra J, Retuerto M, Tatsuoka C, Ghannoum MA, McComsey GA. Dysbiosis in the oral bacterial and fungal microbiome of HIV-infected subjects is associated with clinical and immunologic variables of HIV infection. PLoS One 2018;13:e0200285.


51. Jha AK, Uppal B, Chadha S, Bhalla P, Ghosh R, Aggarwal P, et al. Clinical and microbiological profile of HIV/AIDS cases with diarrhea in North India. J Pathog 2012;2012:971958.


52. Mar Rodríguez M, Pérez D, Javier Chaves F, Esteve E, Marin-Garcia P, Xifra G, et al. Obesity changes the human gut mycobiome. Sci Rep 2015;5:14600.


53. Shah S, Locca A, Dorsett Y, Cantoni C, Ghezzi L, Lin Q, et al. Alterations of the gut mycobiome in patients with MS. EBioMedicine 2021;71:103557.


54. Zhang X, Pan L, Zhang Z, Zhou Y, Jiang H, Ruan B. Analysis of gut mycobiota in first-episode, drug-naïve Chinese patients with schizophrenia: a pilot study. Behav Brain Res 2020;379:112374.

55. Severance EG, Alaedini A, Yang S, Halling M, Gressitt KL, Stallings CR, et al. Gastrointestinal inflammation and associated immune activation in schizophrenia. Schizophr Res 2012;138:48-53.


1. Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct Target Ther 2022;7:1-28.
2. Matijašić M, Meštrović T, Čipčić Paljetak H, Perić M, Barešić A, Verbanac D. Gut microbiota beyond bacteria—mycobiome, virome, archaeome, and eukaryotic parasites in IBD. Int J Mol Sci 2020;21:2668.
3. Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013;14:685-90.
4. Wang H, Wei CX, Min L, Zhu LY. Good or bad: gut bacteria in human health and diseases. Biotechnol Biotechnol Equip 2018;32:1075-80.
5. LeBlanc JG, Milani C, De Giori GS, Sesma F, Van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol 2013;24:160-8.
6. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016;14:e1002533.
7. Tiew PY, Mac Aogain M, Ali NAtBM, Thng KX, Goh K, Lau KJ, et al. The mycobiome in health and disease: emerging concepts, methodologies and challenges. Mycopathologia 2020;185:207-31.
8. Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med 2013;5:1-12.
9. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59-65.
10. Kapitan M, Niemiec MJ, et al. eds. Fungi as part of the microbiota and interactions with intestinal bacteria. In: Rodrigues ML. Fungal physiology and immunopathogenesis. Switzerland; Springer, 2019:265-301.
11. Richard ML and Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 2019;16:33145.
12. Yan Q, Li S, Yan Q, Huo X, Wang C, Wang X, et al. A genomic compendium of cultivated human gut fungi characterizes the gut mycobiome and its relevance to common diseases. Cell 2024;187:2969-89.
13. Bliss JM, Basavegowda KP, Watson WJ, Sheikh AU, Ryan RM. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr Infect Dis J 2008;27:231-5.
14. Nagata R, Nagano H, Ogishima D, Nakamura Y, Hiruma M, Sugita T. Transmission of the major skin microbiota, Malassezia, from mother to neonate. Pediatr Int 2012;54:350-5.
15. Juyal D, Sharma M, Pal S, Rathaur VK, Sharma N. Emergence of non-albicans Candida species in neonatal candidemia. N Am J Med Sci 2013;59:541-5.
16. Ward TL, Dominguez-Bello MG, Heisel T, Al-Ghalith G, Knights D, Gale CA. Development of the human mycobiome over the first month of life and across body sites. MSystems 2018;3:10.1128.
17. Zhang F, Aschenbrenner D, Yoo JY, Zuo T. The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe 2022;3:e969-83.
18. Suhr MJ, Banjara N, Hallen‐Adams HE. Sequence‐based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol 2016;62:209-15.
19. Hallen-Adams H and Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence 2017;8:352–8.
20. Otašević S, Momčilović S, Petrović M, Radulović O, Stojanović N, Arsić-Arsenijević V. The dietary modification and treatment of intestinal Candida overgrowth–a pilot study. J Mycol Med 2018;28:623-7.
21. Strati F, Di Paola M, Stefanini I, Albanese D, Rizzetto L, Lionetti P, et al. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front Microbiol 2016;7:1227.
22. Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut 2017;66:1039-48.
23. Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019;68:654-62.
24. Aminudin NI, Amran NA, Zainal Abidin ZA, Susanti D. Biotransformation of curcumin and structure–activity relationship (SAR) of its analogues: a systematic review. Biocatal Biotransformation 2023;41:1-14.
25. Bishu S, Su EW, Wilkerson ER, Reckley KA, Jones DM, McGeachy MJ, et al. Rheumatoid arthritis patients exhibit impaired Candida albicans-specific Th17 responses. Arthritis Res Ther 2014;16:1-9.
26. Zou R, Wang Y, Duan M, Guo M, Zhang Q, Zheng H. Dysbiosis of gut fungal microbiota in children with autism spectrum disorders. J Autism Dev Disord 2021;51:267-75.
27. Heisel T, Montassier E, Johnson A, Al-Ghalith G, Lin YW, Wei LN, et al. High-fat diet changes fungal microbiomes and interkingdom relationships in the murine gut. mSphere 2017;2:e00351-17.
28. Lapiere A and Richard ML. Bacterial-fungal metabolic interactions within the microbiota and their potential relevance in human health and disease: a short review. Gut Microbes 2022;14:2105610.
29. MacAlpine J, Daniel-Ivad M, Liu Z, Yano J, Revie NM, Todd RT, et al. A small molecule produced by Lactobacillus species blocks Candida albicans filamentation by inhibiting a DYRK1-family kinase. Nat Commun 2021;12:6151.
30. Liu J, Huang TY, Liu G, Ye Y, Soteyome T, Seneviratne G, et al. Microbial interaction between Lactiplantibacillus plantarum and Saccharomyces cerevisiae: transcriptome level mechanism of cell-cell antagonism. Microbiol Spectr 2022;10:e01433-22.
31. Golan-Rozen N, Chefetz B, Ben-Ari J, Geva J, Hadar Y. Transformation of the recalcitrant pharmaceutical compound carbamazepine by Pleurotus ostreatus: role of cytochrome P450 monooxygenase and manganese peroxidase. Environ Sci Technol 2011;45:6800-5.
32. Martínez‐Ramírez JA, Walther G, Peters FT. Studies on drug metabolism by fungi colonizing decomposing human cadavers. Part II: biotransformation of five model drugs by fungi isolated from post‐mortem material. Drug Test Anal 2015;7:265-79.
33. Huang Y, Cao S, Zhang Q, Zhang H, Fan Y, Qiu F, et al. Biological and pharmacological effects of hexahydrocurcumin, a metabolite of curcumin. Arch Biochem Biophys 2018;646:317.
34. Zhang Z, Luo D, Xie J, Lin G, Zhou J, Liu W, et al. Octahydrocurcumin, a final hydrogenated metabolite of curcumin, possesses superior anti-tumor activity through induction of cellular apoptosis. Food Funct 2018;9:2005-14.
35. Zhang J, Guo H, Tian Y, Liu P, Li N, Zhou J, et al. Biotransformation of 20 (S)protopanaxatriol by Mucor spinosus and the cytotoxic structure activity relationships of the transformed products. Phytochemistry 2007;68:2523-30.
36. He X, Zhang B, Cao P, Wang H, Wu S, Wang G, et al. Biotransformation of dihydrocapsaicin by human intestinal fungi and the inhibitory effects of metabolites against LSD1. Heliyon 2022;8:e12325.
37. Zhang M, Zhang J, Wang C, Yan JK, Yi J, Ning J, et al. Biotransformation of 18β-glycyrrhetinic acid by human intestinal fungus Aspergillus niger RG13B1 and the potential anti-inflammatory mechanism of its metabolites. J Agric Food Chem 2022;70:1510415.
38. Wei X, Yao C, He X, Li J, Wang Y, Wang C, et al. Biotransformation of chenodeoxycholic acid by human intestinal fungi and the agonistic effects on FXR. Phytochemistry 2024;224:114162.
39. Bowman MJ, Jordan DB, Vermillion KE, Braker JD, Moon J, Liu ZL. Stereochemistry of furfural reduction by a Saccharomyces cerevisiae aldehyde reductase that contributes to in situ furfural detoxification. Appl Environ Microbiol 2010;76:4926-32.
40. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. PNAS 2012;109:6241-6.
41. Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017;5:1-13.
42. Xie Z and Manichanh C. FunOMIC: pipeline with built-in fungal taxonomic and functional databases for human mycobiome profiling. Comput Struct Biotechnol J 2022;20:3685-94.
43. Langsiri N, Worasilchai N, Irinyi L, Jenjaroenpun P, Wongsurawat T, Luangsa-Ard JJ, et al. Targeted sequencing analysis pipeline for species identification of human pathogenic fungi using long-read nanopore sequencing. IMA fungus 2023;14:18.
44. Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015;18:489-500.
45. Lee EH, Kim H, Koh JH, Cha KH, Lee KK, Kim WU, et al. Dysbiotic but nonpathogenic shift in the fecal mycobiota of patients with rheumatoid arthritis. Gut Microbes 2022;14:2149020.
46. Wang H, Wu H, Li KD, Wang YY, Huang RG, Du YJ, et al. Intestinal fungi and systemic autoimmune diseases. Autoimmun Rev 2023;22:103234.
47. Lin Y, Lau HCH, Liu Y, Kang X, Wang Y, Ting NLN, et al. Altered mycobiota signatures and enriched pathogenic Aspergillus rambellii are associated with colorectal cancer based on multicohort fecal metagenomic analyses. Gastroenterology 2022;163:908-21.
48. Wang T, Fan C, Yao A, Xu X, Zheng G, You Y, et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity 2018;49:504-14.e4
49. Chen Y, Chen Z, Guo R, Chen N, Lu H, Huang S, et al. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis 2011;70:492-8.
50. Mukherjee PK, Chandra J, Retuerto M, Tatsuoka C, Ghannoum MA, McComsey GA. Dysbiosis in the oral bacterial and fungal microbiome of HIV-infected subjects is associated with clinical and immunologic variables of HIV infection. PLoS One 2018;13:e0200285.
51. Jha AK, Uppal B, Chadha S, Bhalla P, Ghosh R, Aggarwal P, et al. Clinical and microbiological profile of HIV/AIDS cases with diarrhea in North India. J Pathog 2012;2012:971958.
52. Mar Rodríguez M, Pérez D, Javier Chaves F, Esteve E, Marin-Garcia P, Xifra G, et al. Obesity changes the human gut mycobiome. Sci Rep 2015;5:14600.
53. Shah S, Locca A, Dorsett Y, Cantoni C, Ghezzi L, Lin Q, et al. Alterations of the gut mycobiome in patients with MS. EBioMedicine 2021;71:103557.
54. Zhang X, Pan L, Zhang Z, Zhou Y, Jiang H, Ruan B. Analysis of gut mycobiota in firstepisode, drug-naïve Chinese patients with schizophrenia: a pilot study. Behav Brain Res 2020;379:112374.
55. Severance EG, Alaedini A, Yang S, Halling M, Gressitt KL, Stallings CR, et al. Gastrointestinal inflammation and associated immune activation in schizophrenia. Schizophr Res 2012;138:4853.