1Department of Laboratory Medicine, Hallym University College of Medicine, Kangdong Sacred Heart Hospital, Seoul, Korea
2Department of Laboratory Medicine, Sahmyook Medical Center, Seoul, Korea
3Department of Laboratory Medicine, Hallym University College of Medicine, Hangang Sacred Heart Hospital, Seoul, Korea
4Department of Laboratory Medicine, Hallym University College of Medicine, Dongtan Sacred Heart Hospital, Hwaseong, Korea
5Department of Laboratory Medicine, Hallym University College of Medicine, Hallym University Sacred Heart Hospital, Anyang, Korea
6Department of Laboratory Medicine, Hallym University College of Medicine, Kangnam Sacred Heart Hospital, Seoul, Korea
*These authors contributed equally to this work.
Correspondence to Jae-Seok Kim, E-mail: jaeseok@kdh.or.kr
Ann Clin Microbiol 2025;28(3):15. https://doi.org/10.5145/ACM.2025.28.3.4
Received on 23 May 2025, Revised on 20 June 2025, Accepted on 30 June 2025, Published on 10 September 2025.
Copyright © Korean Society of Clinical Microbiology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND) (https://creativecommons.org/licenses/by-nc-nd/4.0/).
Background: Methicillin-resistant Staphylococcus aureus (MRSA) is a major pathogen responsible for various clinical infections. The investigation of representative MRSA genomes is important for understanding their molecular epidemiology and genetic evolution, as well as MRSA infections. We characterized the complete genome sequences of representative MRSA clinical strains prevalent in Korea between 2014 and 2017.
Methods: Ten representative clinical MRSA strains were selected based on the Staphylococcal Cassette Chromosome mec (SCCmec) type. Complete genomes were generated via hybrid assembly using long- and short-read sequencing. Analyses of resistance and virulence genes, whole-genome alignment, phylogenetic tree construction, and comparative genome hybridization were performed.
Results: The average chromosomal lengths were 2.916 Mb in SCCmec II (n = 6), 2.920 Mb in SCCmec IV (n = 2), and 2.777 Mb in SCCmec IVA (n = 2). The number of genome coding sequences ranged from 2,713 to 3,026, with an average of 2,946 in SCCmec II, 3,001 in SCCmec IV, and 2,740 in SCCmec IVA. Only the SCCmec IV and spa t008 strains (n = 2) harbored the Panton–Valentine leukocidin gene, which is rarely detected in Korea. The SCCmec IVA strains of ST72 showed a distinct genetic group compared with other representative SCCmec IV strains, as determined by single-nucleotide polymorphism analysis.
Conclusion: In the present study, the complete and gap-filled genome sequences of representative MRSA clones prevalent in Korea were derived and characterized by genome size, virulence, antimicrobial resistance genes, and their evolutionary relationships. Information on these clinical MRSA strains would enhance our understanding of the pathogenicity and molecular epidemiology of Korean MRSA isolates.
Antimicrobial resistance, Methicillin-resistant Staphylococcus aureus, Molecular epidemiology, Staphylococcal protein A, Whole-genome sequencing
Methicillin-resistant Staphylococcus aureus (MRSA) is a leading cause of healthcare- and communityassociated infections and causes significant morbidity and mortality in patients [1]. In Korea, S. aureus is the second most common pathogen causing bloodstream infections, with an incidence of 2.8 cases per 10,000 patient-days among inpatients; more than half of these cases (54.3%) were attributable to MRSA [2]. In the United States, approximately 323,700 MRSA infections and 10,600 associated deaths were reported among hospitalized patients in 2017 [3]. Over the past few decades, the epidemiology of MRSA infections has changed, resulting in severe clinical infections caused by both healthcare-associated MRSA clones and community-associated MRSA (CA-MRSA) clones [4,5]. Understanding the genomic characteristics of the prevalent MRSA strains according to their genetic subtypes is necessary to study MRSA infections and epidemiology.
Whole-genome sequencing (WGS) has revealed the comprehensive and high-resolution genomic characteristics of various bacterial pathogens. Conventional typing methods, such as pulsed-field gel electrophoresis and multilocus sequence typing (MLST), have been successfully used to characterize bacterial strains responsible for disease outbreaks, but offer limited discrimination between closely related strains. WGS has enabled the investigation of pathogens with much greater discriminatory power, resulting in a more comprehensive understanding of the transmission of isolates and their evolutionary relationships [6]. In addition, WGS provides a complete view of antimicrobial resistance genes, virulence factor genes, and other genomic characteristics related to pathogenicity [7,8].
The genome sequence of fragmented DNA contigs with gaps, usually obtained by short-read sequencing technology, has been used to investigate the antimicrobial resistance, epidemiological characteristics, and virulence factors of bacteria. In addition, short-read sequencing data can be further analyzed using referencebased sequence alignment, allowing genome-wide comparisons with other strains with high accuracy [6].
In contrast, the Oxford Nanopore Technologies DNA sequencing platform generates long reads that facilitate complete genome assembly. However, these long reads have relatively higher error rates. Hybrid assembly, which combines short- and long-read sequencing data, using software such as Unicycler [9], can produce complete circular bacterial genomes with high accuracy.
Complete genomes of pathogens provide advantages over fragmented bacterial genomes with gaps because they precisely identify all the genes (i.e., virulence genes and antimicrobial resistance genes) present, and also provide structural information relating to genes, mobile genetic elements, and gene rearrangements. This comprehensive information provides novel structural insights into bacterial genetics [10-12]. In addition, complete genomic sequences enable accurate phylogenetic and evolutionary analyses, including the identification of the horizontal gene transfer of antimicrobial resistance elements [13,14]. Although several complete circular genome sequences of MRSA strains have been published [14-16], comprehensive genomic analyses of MRSA clinical isolates from Korea remain scarce, with only approximately five MRSA isolates to date. The paucity of genomic representation limits our understanding of the molecular characteristics of Korean MRSA strains.
The present study was conducted to address this limitation by performing complete genome sequencing of MRSA clinical isolates from Korea. We selected ten representative MRSA strains from blood specimens collected at a university hospital according to the molecular genotypes of their staphylococcal cassette chromosome mec (SCCmec) and staphylococcal protein A (spa) type. To reveal the characteristics of these MRSA strains, complete circular genomes were derived using hybrid assembly. The genome sizes of representative MRSA clones in Korea and the characteristics of their virulence factors and antimicrobial resistance were investigated herein. Phylogenetic analyses were performed to compare the genomic characteristics of Korean MRSA strains with those of representative S. aureus strains from other countries.
Ten MRSA clinical isolates obtained from the blood cultures of patients presenting at Kangdong Sacred Heart Hospital, Seoul, Korea, between 2014 and 2017, were selected according to their SCCmec and spa types (Table 1). We performed SCCmec typing on all MRSA blood isolates at our hospital during this period. Among these isolates, representative strains were selected to reflect the major genotypes prevalent in Korea: SCCmec II (n = 6) with spa t002, t2460, and t9353; SCCmec IV (n = 2) with spa t008; SCCmec IVA (n = 2) with spa t324. These combinations are commonly associated with the predominant hospital- and communityassociated MRSA clones in Korea. The strains were also identified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Bruker Microflex LT; Bruker Daltonics GmbH), and MRSA was confirmed by polymerase chain reaction amplification of mecA [15] and cefoxitin disk diffusion tests. The SCCmec [17] and spa types [18] were determined for each MRSA isolate.
Table 1. Summary metrics of the assembled (circular) and annotated genomes of methicillin-resistant Staphylococcus aureus strains
| Strain | HL18888 | HL20835 | HL17064 | HL18807 | HL18883 | HL21008 | HL17078 | HL18380 | HL16278 | HL18840 |
|---|---|---|---|---|---|---|---|---|---|---|
| SCCmec type | II | II | II | II | II | II | IV | IV | IVA | IVA |
| spa type | t002 | t002 | t2460 | t2460 | t9353 | t9353 | t008 | t008 | t324 | t324 |
| Isolation year | 2016 | 2017 | 2015 | 2016 | 2016 | 2017 | 2015 | 2016 | 2014 | 2016 |
| Completeness (%)* | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Chromosomes | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Total length (bp) | 2,893,276 | 2,913,324 | 2,931,169 | 2,903,929 | 2,904,018 | 2,947,630 | 2,922,932 | 2,917,999 | 2,757,377 | 2,796,963 |
| GC (%) | 32.89 | 32.91 | 32.98 | 32.92 | 32.92 | 32.93 | 32.81 | 32.8 | 32.82 | 32.85 |
| CDS | 2,923 | 2,970 | 2,974 | 2,905 | 2,906 | 3,000 | 3,026 | 2,975 | 2,713 | 2,766 |
| rRNAs | 16 | 16 | 16 | 16 | 16 | 16 | 19 | 19 | 16 | 19 |
| tRNAs | 59 | 59 | 59 | 59 | 59 | 59 | 59 | 59 | 59 | 59 |
| ncRNAs | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| GenBank No. | CP080548.1 | CP080566.1 | CP080560.1 | CP080552.1 | CP080550.1 | CP080562.1 | CP080556.1 | CP080553.1 | CP080564.1 | CP080551.1 |
| Plasmids | 1 | 1 | 1 | 0 | 0 | 1 | 3 | 2 | 1 | 0 |
| Plasmid length (bp) | 24,653 | 24,653 | 25,109 | 25,109 | 42,253/19,845/3,125 | 23,058/6,118 | 3,332 | |||
| GC (%) | 28.5 | 28.5 | 29 | 29 | 28.5/29/28.5 | 30.5/28.5 | 29.5 | |||
| GenBank No. | CP080549.1 | CP080567.1 | CP080561.1 | CP080563.1 | CP080557.1/ CP080559.1/ CP080559.1 | CP080554.1/ CP080555.1 | CP080565.1 | |||
| Genome assembly No. | ASM1955137v1 | ASM1955141v1 | ASM1955094v1 | ASM1955131v1 | ASM1955103v1 | ASM1955109v1 | ASM1955117v1 | ASM1955123v1 | ASM1955135v1 | ASM1955087v1 |
The table was created with analysis of genome statistics of Fasta sequences.
aCompleteness of genome assembly was analyzed by BUSCO v.5.3.2 with bacteria_odb10 lineage dataset.
Abbreviations: SCCmec, staphylococcal cassette chromosome mec; spa, staphylococcal protein A; bP, base pairs; GC, guanine and cytosine; CDS, coding sequences or protein coding genes; BUSCO, benchmarking universal single-copy ortholog.
The MRSA strains were cultured on 5% sheep blood agar plates at 37°C under 5% CO2 for 16–18 h. Genomic DNA was extracted using QIAamp® DNA Mini Kit (Qiagen), and the DNA concentration was measured with a Quantus™ Fluorometer (Quantus Inc.).
For short-read sequencing, genome libraries were prepared using Nextera DNA Flex Library Prep Kit (Illumina Inc.). The size and concentration of DNA libraries were measured using a TapeStation 4200 capillary electrophoresis platform (Agilent Inc.). Paired-end libraries were sequenced using the Illumina MiSeq platform [15].
For long-read sequencing, genome libraries were prepared using a ligation sequencing kit (SQK-LSK109; Oxford Nanopore Technologies) and sequenced using a MinION flow cell (FLO-MIN106D; R9; Oxford Nanopore Technologies) controlled by the MinKNOW software (v.20.10.3).
Genomes were assembled with Unicycler (v.0.4.8) [9] using MiSeq paired-end sequences and MinION sequences. Genomes were annotated using the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP) [19]. Complete genome sequence data for MRSA strains have been deposited in the NCBI database with BioProject PRJNA693997 (Dataset 1).
Mauve analysis using MegAlign Pro (DNASTAR v.17.0.2.1) was performed to align the MRSA circular genome sequences and identify the structural characteristics of the genome.
With the genome of the study isolates and the previously reported representative strains, single-nucleotide polymorphism (SNP)-based genome alignment was performed using Snippy (v.4.6.0) and Gubbins (v.3.4) was used to exclude recombination. The N315 complete genome was selected as the reference genome. Based on recombinant-free alignments, phylogenetic analysis was performed using RAxML-NG (v.1.2.2) [20] for the maximum-likelihood tree. A phylogenetic tree was constructed using FigTree software (v.1.4.4). Core- and whole-genome MLST analyses were performed using ChewBBACA software (v.3.3.10). Newick-formatted tree files of MSTreeV2, a minimal spanning tree method, were generated using Grapetree (v.1.5.0), and trees were drawn using FigTree.
Maps of the constructed MRSA genomes were generated. A visual comparison of the ten MRSA WGS datasets was performed using the CGView Comparison Tool (CCT; v.2.0.3) [21]. Clusters of orthologous gene (COG) functional categories were assigned based on the NCBI COG database (https://www.ncbi.nlm. nih.gov/research/cog) and are displayed using different colors. Roary (v.3.13.0) was used herein to determine the presence and absence of genes and the number of core genes [22].
The MicroScan WalkAway 96 Plus system (Beckman Coulter) was used to determine the phenotypic antimicrobial susceptibility testing (AST) of each MRSA strain. Using the MRSA genome sequences, the presence of antimicrobial resistance and virulence genes was identified using ResFinder (v.4.4.1) and VirulenceFinder (v.2.0.3) from the Center for Genomic Epidemiology server. Both Resfinder and VirulenceFinder were set to a default threshold ID of 90% and minimum length of 60%.
Each of the ten MRSA strains possessed a single circular chromosome, ranging from 2,757,377 to 2,947,630 base pairs (bp), with guanine and cytosine (GC) content between 32.80% and 32.98% (average 32.88%) (Table 1). The average chromosomal lengths were 2.916 Mb for SCCmec II (n = 6), 2.920 Mb for SCCmec IV (n = 2), and 2.777 Mb for SCCmec IVA (n = 2).
The SCCmec II and SCCmec IVA strains carried either no plasmids or a single plasmid, whereas the spa t008 SCCmec IV strain carried two or three plasmids (Table 1). Notably, two SCCmec II strains of spa t002 harbored plasmids of identical size (24,653 bp), whereas one t2460 strain and one t9353 strain also carried plasmids of the same size (25,109 bp), regardless of the isolation year.
The MRSA genome contains 2,713–3,026 protein-coding genes and 79–82 non-coding genes. The number of coding sequences (CDS) ranged from 2,713 to 3,026, with an average of 2,946 in SCCmec II, 3,001 in SCCmec IV, and 2,740 in SCCmec IVA. Roary pan-genome analysis identified 3,661 total genes, including 2,204 core genes (present in ≥ 99% of strains), 0 soft-core genes (95%-98%), 1,151 shell genes (15%-94%), and 306 cloud genes (< 15%). Information pertaining to the gene content of the MRSA strains is listed in supplementary Table 1 in the online-only Data Supplement. The GC content of the genomes showed minimal variation among strains (range: 32.80–32.98, average: 32.88).
Ten MRSA strains represented five spa types (t002, t008, t324, t2460, and t9353), with two isolates per spa type. The spa strains t002, t2460, and t9353 were associated with SCCmec II (n = 6), t008 with SCCmec IV (n = 2), and t324 with SCCmec IVA (n = 2). All SCCmec II isolates belonged to MLST ST5, while two SCCmec IV strains belonged to ST5 and ST5863, and two SCCmec IVA strains belonged to ST72.
Mauve analysis of the complete genomes revealed high levels of synteny between these representative MRSA genomes (Fig. 1), although structural variations, such as intraspecific recombination, were noted. Gene order in the proximal and distal regions was conserved; however, structural differences, such as recombination, inversion, and deletions, occurred mainly between ~0.9 Mb and ~2.2 Mb. A large genomic inversion (~0.8 Mb) was observed in strain HL17064.
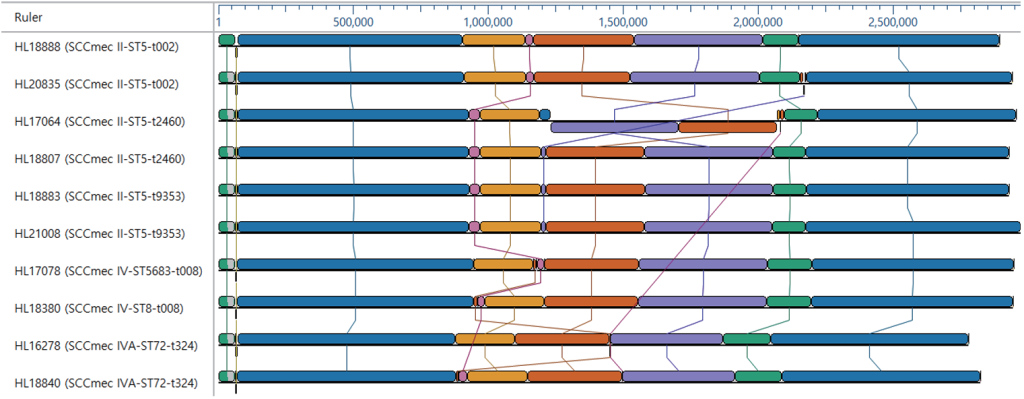
Fig. 1. Genome alignments of MRSA strains using multiple alignment of conserved genomic sequences (MAUVE). Pairwise comparisons of MRSA genome sequences revealed a large genomic inversion in strain HL17064. MRSA: methicillin-resistant Staphylococcus aureus
Phylogenetic tree analysis based on SNPs of the genome showed that strains grouped by SCCmec type formed three distinct clades (Fig. 2). Korean MRSA strains of the same SCCmec type clustered closely. The SCCmec II strain, ST5 (CC5)-t002/t2460/t9353, is closely related to representative ST5 strains, such as Mu50, N315, and JH1. The SCCmec IV strain, ST8/ST5863 (CC8)-t008, clustered with the representative ST8 strains TW20, Newman, USA300_FPR3757, COL, and NCTC 8353. Interestingly, the Korean SCCmec IVa strain ST72 (CC8)-t324 formed a unique phylogenetic clade distinct from the CC8 representative strains.
For the Korean SCCmec IVa strain of ST72-t324, additional analyses with cgMLST and wgMLST, the allele-based genotyping methods using fewer loci (2,009 and 3,567 loci), also showed findings compatible with those of Snippy (112,574 loci), the whole-genome SNP-based analysis (Fig. 3).
Circular genome maps were generated using the largest genome (HL21008) as reference (Fig. 4). Genes in the SCCmec II strains showed ≥ 98% identity, while the SCCmec IV strains displayed 94%–98% similarity in some genomic elements. A lack of genetic elements was observed in specific SCCmec and spa types. For instance, the SCCmec IV and IVA strains (inner four circles) lacked phenol-soluble modulin-mec (psmmec), mecI, and part of mecR in the SCCmec region (~37–39 kb region) of the proximal part of the MRSA genome (supplementary Fig. 1 in the online-only Data Supplement).
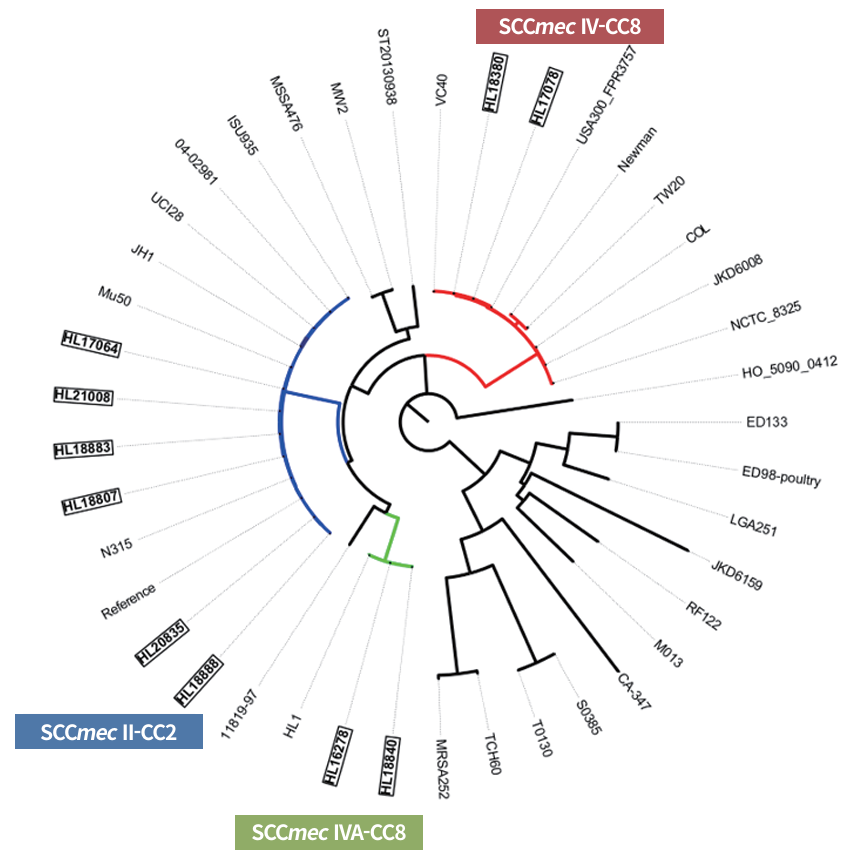
Fig. 2. SNP-based phylogenetic tree with Snippy, Gubbins, and RAxML-NG analysis of the representative S. aureus strains, illustrating relationships between major clones. The Korean clinical MRSA strains are indicated by bolded rectangles. SCCmec II strains from Korea clustered closely with other representative SCCmec II strains. In contrast, SCCmec IVA strains of CC8 (HL16278 and HL18840) formed a distinct clade, separate from other SCCmec IV reference strains of CC8. A total of 112,574 SNP loci were included in the analysis. SNP: single-nucleotide polymorphism; MRSA: methicillin-resistant Staphylococcus aureus; SCCmec: staphylococcal cassette chromosome mec
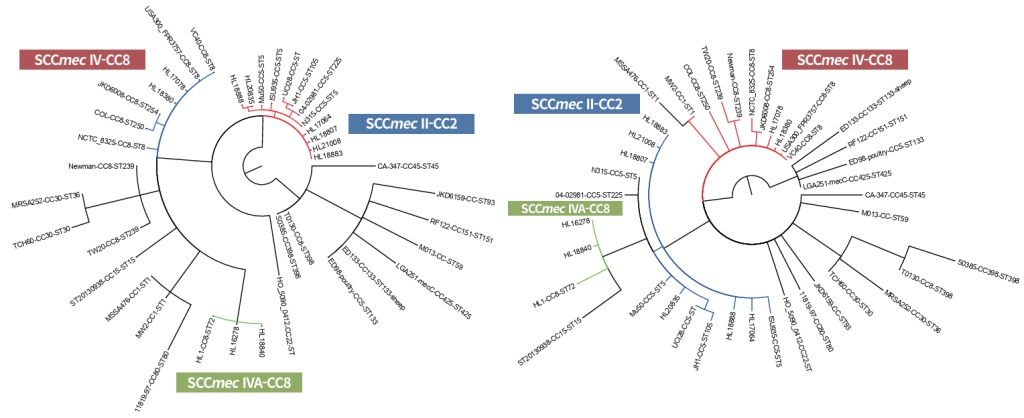
Fig. 3. Minimal spanning tree (MSTreeV2) based on cgMLST (left) and wgMLST (right) allelic profiles of the representative S. aureus strains, illustrating relationships between major clones. The Korean clinical MRSA strains are indicated by bolded rectangles. SCCmec II strains from Korea clustered closely with other representative SCCmec II strains. In contrast, SCCmec IVA strains of CC8 (HL16278 and HL18840) formed a distinct clade, separate from other SCCmec IV reference strains of CC8 in both allelic analysis of cgMLST and wgMLST, which is consistent with the findings of SNP-based phylogenetic tree analysis. A total of 2,009 cgMLST loci and 3,567 wgMLST loci were included in this analysis. MLST: multilocus sequence typing; SCCmec: staphylococcal cassette chromosome mec; SNP: single-nucleotide polymorphism
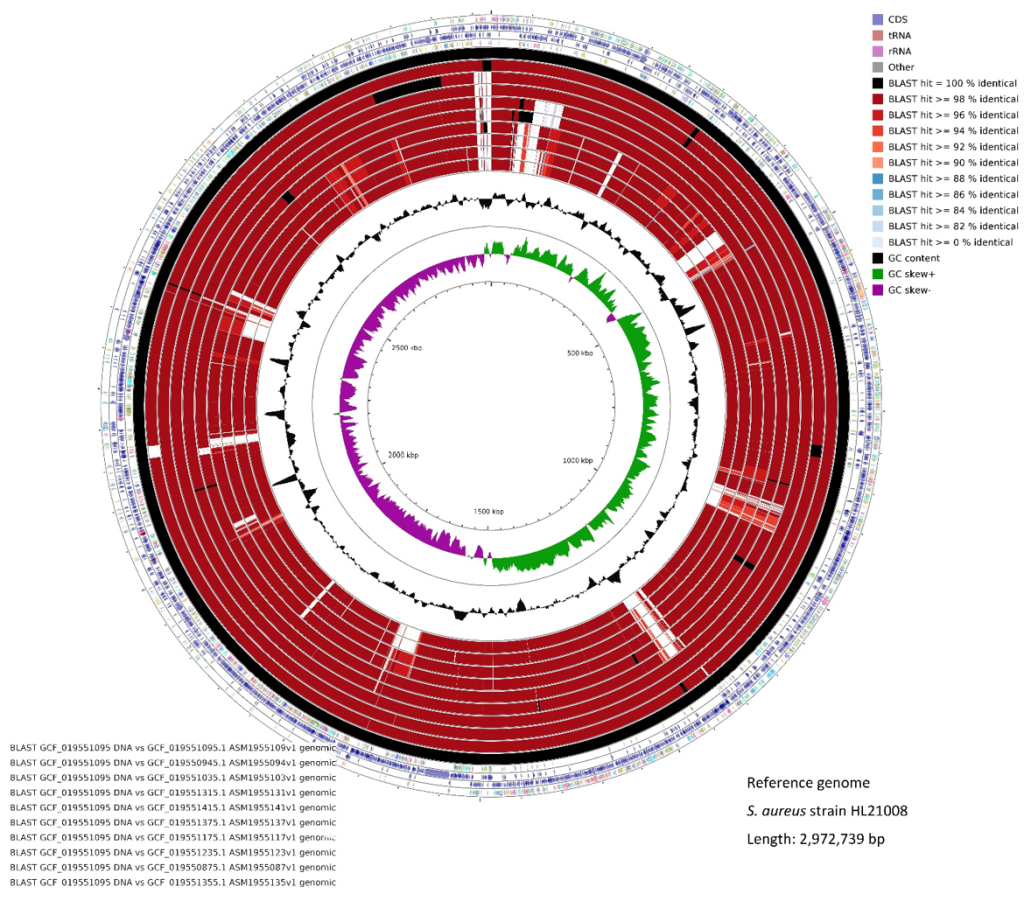
Fig. 4. Circular visualization of MRSA complete genomes with HL21008 as a reference strain. The inner genome rings showed 100% identity (black), >= 98% (dark magenta) and >= 96% (red) for these reference isolates, reflecting similarity percentages determined by BLAST hit. Black peaks indicate GC content, and the innermost green and purple peaks denote positive and negative GC skews, respectively. MRSA: methicillin-resistant Staphylococcus aureus
The mecA gene was detected in all the MRSA strains using ResFinder. The blaZ beta-lactamase gene was found in strains spa t002, t008, and t324, but was absent in the t2460 and t9353 strains of SCCmec II (Table 2). All isolates were phenotypically resistant to beta-lactam antibiotics ampicillin, oxacillin, and penicillin.
There was full concordance between the phenotypic AST and genotypic AST for gentamicin (aac(6′)aph(2”)), ciprofloxacin (grlA and gyrA mutations), clindamycin (erm(A)), erythromycin (erm(A) or msr(A)), fusidic acid (fusC or fusA (L461K)), and mupirocin (ileS (V588F) or mupA).
Table 2. Phenotypic and genotypic antimicrobial susceptibility test (AST) results of methicillin-resistant Staphylococcus aureus strains
| Strain | HL18888 | HL20835 | HL17064 | HL18807 | HL18883 | HL21008 | HL17078 | HL18380 | HL16278 | HL18840 |
|---|---|---|---|---|---|---|---|---|---|---|
| SCCmec type | II | II | II | II | II | II | IV | IV | IVA | IVA |
| spa type | t002 | t002 | t2460 | t2460 | t9353 | t9353 | t008 | t008 | t324 | t324 |
| Isolation year | 2016 | 2017 | 2015 | 2016 | 2016 | 2017 | 2015 | 2016 | 2014 | 2016 |
| MLST ST | ST5 | ST5 | ST5 | ST5 | ST5 | ST5 | ST5863 | ST8 | ST72 | ST72 |
| MLST CC | CC5 | CC5 | CC5 | CC5 | CC5 | CC5 | CC8 | CC8 | CC8 | CC8 |
| Phenotypic resistance profile* | OX, PEN, AMP, AMC, AZI, CD#, CIP, ERY, FA#, IMI, LVX, MXF | OX, PEN, AMP, AMC, AZI, CD, CIP, ERY, GM, IMI, LVX, MXF | OX, PEN, AMP, AMC, AZI, CD, CIP, ERY, FA, FOS, GM, IMI, LVX, MXF, TE | OX, PEN, AMP, AMC, AZI, CD, CIP, ERY, FA, FOS, IMI, LVX, MUP, MXF, TE | OX, PEN, AMP, AMC, AZI, CD, CIP, ERY, FA, FOS, IMI, LVX, MXF, TE | OX, PEN, AMP, AMC, AZI, CD, CIP, ERY, FA, FOS, GM, IMI, LVX, MXF | OX, PEN, AMP, AMC, AZI, CIP, ERY, GM, IMI, LVX, MUP, MXF | OX, PEN, AMP, AMC, AZI, CIP, ERY, IMI, LVX, MXF, RIF, | OX, PEN, AMP, AMC, IMI | OX, PEN, AMP, AMC, IMI |
| Genotypic antimicrobial resistance** | mecA, blaZ, aadD, erm(A), tetM, fusC | mecA, blaZ, aac(6′)-aph(2”), erm(A), tetM | mecA, aac(6′)-aph(2”), erm(A), tetM | mecA, erm(A), tetM | mecA, erm(A), tetM | mecA, aac(6′)-aph(2”), erm(A), tetM | mecA, blaZ, aac(6′)-aph(2”), msr(A), mph(C), mupA | mecA, blaZ, aph(3′)-III, msr(A), mph© | mecA, blaZ, aadD | mecA, blaZ |
| Mutations in AMR genes$ | grlA (S80F, E84K) + gyrA (S84L, S85P) | grlA (S80F) + gyrA (S84L) | grlA (S80F) + gyrA (S84L), fusA (L461K) | grlA (S80F) + gyrA (S84L), fusA (L461K), ileS (V588F) | grlA (S80F) + gyrA (S84L), fusA (L461K) | grlA (S80F) + gyrA (S84L), fusA (L461K) | grlA (S80Y) + gyrA (S84L) | grlA (S80Y) + gyrA (S84L) | mecA, blaZ, aadD | mecA, blaZ |
aPhenotypic AST was performed using MicroScan WalkAway 96 plus system.as part of the routine clinical practice: bAntimicrobial resistance genes were identified using ResFinder 4.3.3 (≥90% sequence similarity with ≥60% minimum identity length); cHL18888 strain showed intermediate resistance to CD encoded by erm(A) and FA encoded by fusC gene; d( ) within parentheses detected mutations are indicated.
Abbreviations: SCCmec, staphylococcal cassette chromosome mec; spa, staphylococcal protein A; MLST, multilocus sequence type; ST, sequence type; CC, clonal complex; AMR, antimicrobial resistance; AMP, ampicillin; AMC, Amoxicillin/ K Clavulanate (β-lactam/ β-lactamase inhibitor combination); AZI, azithromycin; CD, clindamycin; CIP, ciprofloxacin; ERY, erythromycin; FA, fusidic acid; FOS, fosfomycin; GM, gentamicin; IMI, imipenem (β-lactam antibiotic of subgroup carbapenem); LVX, levofloxacin; MUP, mupirocin; MXF, moxifloxacin; OX, oxacillin; PEN, penicillin; RIF, rifampin; TE, tetracycline.
No antimicrobial resistance genes were detected for sulfaomethoxazole-trimethoprim, fosfomycin, vancomycin, teicoplanin, quinupristin-dalfopristin, linezolid, or chloramphenicol, and all isolates were susceptible to these antimicrobial agents (supplementary Table 2 in the online-only Data Supplement).
A total of 26 virulence genes were identified by the VirulenceFinder (Table 3). All isolates carried aureolysin (aur), gamma-hemolysin chain II precursor (hlgA), gamma-hemolysin B precursor (hlgB), gamma-hemolysin component C (hlgC), leukocidin D component (lukD), leukocidin E component (lukE), serine protease splA (splA), or serine protease splB (splB) genes. Among the SCCmec II strains, toxic shock syndrome toxin-1 (tst) was detected in all but one strain (HL17064; spa t2460). No tst genes were found in the SCCmec IV and IVA strains.
The lukF-PV and lukS-PV genes, encoding Panton–Valentine leukocidin (PVL), and the ACME gene, encoding the arginine catabolic mobile element (ACME), were observed only in the two SCCmec type IV spa t008 strains (HL17078 and HL18380). The SCCmec IV strains also showed unique virulence profiles, including enterotoxin K (sek), enterotoxin Q (seq), and serine protease splE (splE), in contrast to the SCCmec II and IVA strains.
Table 3. Virulence gene profiles of methicillin-resistant Staphylococcus aureus strains
| Strain | HL18888 | HL20835 | HL17064 | HL18807 | HL18883 | HL21008 | HL17078 | HL18380 | HL16278 | HL18840 |
|---|---|---|---|---|---|---|---|---|---|---|
| SCCmec type | II | II | II | II | II | II | IV | IV | IVA | IVA |
| spa type | t002 | t002 | t2460 | t2460 | t9353 | t9353 | t008 | t008 | t324 | t324 |
| Isolation year | 2016 | 2017 | 2015 | 2016 | 2016 | 2017 | 2015 | 2016 | 2014 | 2016 |
| MLST ST | ST5 | ST5 | ST5 | ST5 | ST5 | ST5 | ST5863 | ST8 | ST72 | ST72 |
| MLST CC | CC5 | CC5 | CC5 | CC5 | CC5 | CC5 | CC8 | CC8 | CC8 | CC8 |
| Arginine catabolic mobile element | – | – | – | – | – | – | ACME | ACME | – | – |
| Aureolysin | aur | aur | aur | aur | aur | aur | aur | aur | aur | aur |
| Gamma-hemolysin chain II precursor | hlgA | hlgA | hlgA | hlgA | hlgA | hlgA | hlgA | hlgA | hlgA | hlgA |
| Gamma-hemolysin component B precursor | hlgB | hlgB | hlgB | hlgB | hlgB | hlgB | hlgB | hlgB | hlgB | hlgB |
| Gamma-hemolysin component C | hlgC | hlgC | hlgC | hlgC | hlgC | hlgC | hlgC | hlgC | hlgC | hlgC |
| Leukocidin D component | lukD | lukD | lukD | lukD | lukD | lukD | lukD | lukD | lukD | lukD |
| Leukocidin E component | lukE | lukE | lukE | lukE | lukE | lukE | lukE | lukE | lukE | lukE |
| Leukocidin F component | – | – | – | – | – | – | lukF-PV | lukF-PV | – | – |
| Leukocidin S component | – | – | – | – | – | – | lukS-PV | lukS-PV | – | – |
| Staphylokinase | sak | sak | – | – | – | – | sak | sak | sak | sak |
| Staphylococcal complement inhibitor | scn | scn | – | – | – | – | scn | scn | scn | scn |
| Enterotoxin C | – | sec | – | sec | sec | sec | – | – | – | – |
| Enterotoxin K | – | – | – | – | – | – | sek | sek | – | – |
| Enterotoxin Q | – | – | – | – | – | – | seq | seq | – | – |
| Enterotoxin G | seg | seg | seg | seg | seg | seg | – | – | seg | seg |
| Enterotoxin I | sei | sei | sei | sei | sei | sei | – | – | sei | sei |
| Enterotoxin L | – | sel | – | sel | sel | sel | – | – | – | |
| Enterotoxin M | sem | sem | sem | sem | sem | sem | – | – | sem | sem |
| Enterotoxin N | sen | sen | sen | sen | sen | sen | – | – | sen | sen |
| Enterotoxin O | seo | seo | seo | seo | seo | seo | – | – | seo | seo |
| Enterotoxin P | sep | sep | – | – | – | – | – | – | – | – |
| Enterotoxin U | seu | seu | seu | seu | seu | seu | – | – | seu | seu |
| Serine protease splA | splA | splA | splA | splA | splA | splA | splA | splA | splA | splA |
| Serine protease splB | splB | splB | splB | splB | splB | splB | splB | splB | splB | splB |
| Serine protease splE | – | – | – | – | – | – | splE | splE | – | – |
| Toxic shock syndrome toxin-1 | tst | tst | – | tst | tst | tst | – | – | – | – |
Abbreviations: SCCmec, staphylococcal cassette chromosome mec; spa, staphylococcal protein A; MLST, multilocus sequence type; ST, sequence type; CC, clonal complex.
In the present study, the genomes of 10 clinical MRSA isolates were sequenced using short-read-based MiSeq and long-read-based MinION platforms, and hybrid assembly was performed to assemble the complete MRSA genome sequences and understand the genome structures of representative Korean MRSA strains. Furthermore, WGS analysis of these MRSA strains provided a comprehensive overview of their genetic characteristics (such as genome size, CDS number, and GC content), highlighted differences in virulence factors and antimicrobial resistance genes, and identified molecular epidemiological markers corresponding to the SCCmec and/or spa types.
Investigations of complete bacterial genome sequences have been conducted to understand the full complement of genetic elements and the genome structure of major pathogens, such as S. aureus, E. coli, and Helicobacter pylori [10,12,23]. The availability of whole-genome sequences not only provides information on genetic variations and pathogenicity factors but is also valuable for the investigation of metabolic networks, drug development, and evolution [24-26]. In addition, the complete genome sequences of pathogens are the basis for mutant analysis [26,27], metabolic system analysis [24], and precise comparative genomic analysis [15].
We obtained the complete genomes of 2,038 S. aureus strains isolated worldwide from the NCBI genome database. Our search of this database, using the filters of “Korea” or “Seoul” (https://www.ncbi.nlm.nih.gov/ datasets/genome/?taxon=1280&assembly_level=3:3. Accessed 2025-05-08), revealed only five human MRSA isolates, two methicillin-susceptible Staphylococcus aureus (MSSA) isolates, and four Bos taurus (cattle) MSSA isolates from Korea, excluding the MRSA genome sequences submitted by our group. Four of the five human MRSA strains were isolated in 2011 and 2013.
The genome sizes of the MRSA strains obtained herein ranged from 2.7 to 2.9 Mb, with an average GC content of 32.88% (Table 1), which is consistent with previous published data. Genome size variation probably reflects differences in the presence or absence of mobile genetic elements, virulence factors, and other accessory genetic elements [8,15,23]. The average MRSA chromosomal lengths observed herein were 2.916 Mb in SCCmec II, 2.920 Mb in SCCmec IV, and 2.777 Mb in SCCmec IVA, respectively. SCCmec IVA-ST72-spa t324, a prevalent community-associated MRSA lineage, tends to have a smaller genome than healthcare-associated (i.e., SCCmec II) and community-associated strains (i.e., SCCmec IV). This was because of the smaller size of the SCC regions and the absence of several regions throughout the genome, as indicated by the circular map generated following CGH analysis (Fig. 3).
Genome analysis of global ST72 isolates showed that the ST72 MRSA and MSSA lineages have evolved differently in Asia [8]. Phylogenetic analysis with SNPs and recombinant-free alignment showed that MLST ST72 (CC8) MRSA, a Korean CA-MRSA lineage, was placed in a different clade from other representative CC8 strains (Fig. 2). HL16278, HL18840, and HL1 of SCCmec IVA-ST72 (CC8) were genetically distinct from other SCCmec IV strains of CC8 (i.e., HL17078, HL18380, USA300_FPR3757, and TW20) despite sharing SCCmec elements and four alleles of the MLST genes ST72 (1-4-1-8-4-4-3) and ST8 (3-3-1-1-44-3). These ST72 strains of SCCmec IVA were close to strain11819-97 (SCCmec IVA-ST80); however, the MLST profiles of ST72 (1-4-1-8-4-4-3) and ST80 (1-3-1-14-11-51-10) shared only two alleles. The differences in the genetic background of MLST and SNPs analyzed by WGS suggested that ST72 (CC8) evolved from a genetically distinct lineage from the CC8 strains of other CA-MRSA representative strains, such as the USA300 clone, irrespective of their similarity to the ST and CC types.
Complete genome sequencing, SCCmec typing, and MLST analysis suggest that this successful CAMRSA lineage may have evolved independently from other strains with similar SCCmec types, highlighting the importance of genome studies for epidemiological surveillance. Further studies on bacterial genomes are needed to understand the epidemiology and molecular characteristics of the successful CA-MRSA ST72 lineage in Korea and other neighboring countries.
Plasmid analysis revealed that most MRSA strains possessed a single or no plasmid, whereas SCCmec IV strains (n = 2) harbored two or three plasmids. Small plasmids (< 10 kb) were found only in the SCCmec IV and IVA strains (n = 3; 3,125 bp, 6,118 bp, and 3,332 bp), whereas the SCCmec II strains all had a plasmid > 25 kb in size. The mechanism of replication of plasmids differs based on their size: plasmids smaller than 10 kb generally use the rolling circle mechanism, whereas plasmids larger than 14.5 kb replicate using the thetamode [28]. The biological significance of plasmid size variations among strains remains to be elucidated.
Whole-genome variability using comparative genome analysis was determined by Roary analysis of core and accessory genes. The entire gene pool comprised of 3,661 genes, of which 2,204 were core genes and 1,457 were accessory genes. Large core gene numbers may reflect a high degree of conservation of genes required for the maintenance of essential metabolic mechanisms, virulence, and antimicrobial resistance. In the present study, the presence or absence of genes was used to characterize the MRSA strains (supplementary Table 1 in the online-only Data Supplement); however, further studies with more MRSA strains are needed to understand the general gene pools and genetic plasticity of MRSA in Korea.
For phylogenetic analysis of various representative MRSA strains, we used the Snippy, Gubbins, and RAxML-NG tools. Whole-genome SNP-based and phylogenetic analyses revealed that the MRSA strains clustered into three distinct clades according to their SCCmec types, and into very close clades according to their spa types (Fig. 2). Interestingly, the Korean SCCmec IV and SCCmec IVA strains with the same clonal complex clustered into distinct clades, indicating genomic divergence (Fig. 4 and supplementary Fig. 1 in the online-only Data Supplement).
Single-nucleotide polymorphism-based analysis using Snippy provides the highest resolution for phylogenetic reconstruction by identifying single-nucleotide differences in the core genome relative to the reference genome. By contrast, cgMLST and wgMLST offer allele-based typing approaches that are portable and scalable for large-scale surveillance. While cgMLST focuses on conserved core loci, wgMLST includes both core and accessory genes, providing broader genome coverage but potentially more missing data. We analyzed these methods to provide complementary insights into the genetic relatedness and evolutionary relationships of S. aureus strains.
Studies have shown that a high level of synteny (gene order) is retained among different staphylococcal strains, but intraspecific homologous recombination occasionally occurs [13,29]. In the present study, complete whole genomes allowed Mauve analysis to indicate a relatively well-conserved synteny among most MRSA strains (Fig. 1), although evidence of intraspecific recombination and the presence or absence of accessory genes were observed, particularly between the 0.9 Mb and 2.2 Mb regions of MRSA genomes. The findings obtained herein indicate that frequent structural variations occur primarily in specific genomic regions, as revealed by analysis of complete MRSA genomes. Interestingly, a large genomic inversion was observed in the HL17064 strain. Although substantial information on virulence, epidemiological markers, and resistance genes can be obtained using short-read sequencing[1], complete genome assembly is required to fill the gap in contigs for the identification of precise structural variations and synteny.
Comparative genome hybridization analysis showed genetic homology among the MRSA strains (Fig. 4) and the absence of some genetic elements (i.e., lack of phenol-soluble modulin-mec [psm–mec], mecI, and part of mecR) near the SCCmec regions, especially in the SCCmec IV and IVA strains (supplementary Fig. 1 in the online-only Data Supplement), is a well-known characteristic of SCCmec type IV and IVA strains [30]. We also observed the absence of the cst operon (cstB and cstR), an essential element for sulfide detoxification [31,32] and an erythromycin-resistance gene (ermA) adjacent to the SCCmec region (data not shown). The absence of specific genes near the SCCmec regions in the SCCmec IV and IVA strains suggests evolutionary events that could affect their pathogenicity, adaptation to hospital environments, and antimicrobial-resistance phenotypes [30].
The phenotypic and genotypic AST results of the studied isolates showed 100% concordance with the antimicrobials tested (Table 2).
The MRSA strains were analyzed for the presence of genes encoding 26 different virulence factors using VirulenceFinder (Table 3). All MRSA strains possessed aur, hlgA, hlgB, hlgC, lukD, lukE, splA, and splB, regardless of the SCCmec or spa type, suggesting a core set of virulence factors among the MRSA clinical isolates. The gamma-hemolysin (hlgAB, hlgCB) locus is present in ~99% of sequenced S. aureus genomes, whereas the leukocidin (lukED) locus is present in ~70% of S. aureus isolates and is conserved in a lineagespecific manner [33]. Both SCCmec IV spa t008 strains harbored PVL- and ACME-encoding genes. Such strains are infrequently isolated in Korea [34,35] and exhibit virulence gene profiles that are different from those of the SCCmec II and IVA strains, indicating distinct pathogenic and genomic characteristics. The SCCmec IV strains, in contrast to SCCmec IVA, showed distinct genomic characteristics, suggesting different origins from other countries. The toxic shock syndrome toxin-1 gene (tst) was associated with five of the six SCCmec II strains, but not with the SCCmec IV or IVA strains, suggesting differences in the pathogenic potential of the MRSA strains according to their SCCmec types [36].
Complete genome sequences could provide insight into the regulation of antimicrobial resistance and virulence mechanisms, and identify networks of genes/operons that are coordinately expressed [26], especially for experiments with specific reference or clinical isolates. In addition, reference genomes are necessary for RNA sequencing to obtain a global view of gene expression [37,38]. Such transcriptomic analyses can be useful for investigating bacterial responses to different environmental stresses, such as antimicrobial agents, and for developing novel methods to inhibit MRSA infections [38]. We believe that the complete genomes of Korean MRSA clinical strains would be useful for the investigation of MRSA antimicrobial resistance, gene function, and gene regulation in RNA-seq studies of these clinical isolates.
Although the aim of the present study was to determine the complete genomes of the prevalent Korean MRSA strains, only a limited number of isolates from a single hospital were analyzed. Follow-up studies on a broader and more diverse collection of MRSA strains are required to fully explore the genomic diversity of clinical MRSA isolates.
Complete circular bacterial genome sequences have advantages over fragmented genomes with gaps, as they provide precise genetic information, intact genomic structures, and reference templates for geneexpression studies, such as RNAseq. These complete genomes of representative clinical MRSA strains provide comprehensive and detailed genomic knowledge and are essential for accurate epidemiological and evolutionary studies on S. aureus.
The following supplementary materials are available on the journal’s website:
• Supplementary Table 1. The list of genes in the MRSA study isolates. The number 1 means presence and number 0 means absence of the gene
• Supplementary Table 2. Antimicrobial susceptibility of MRSA strains in this study
• Supplementary Figure 1. The mecA and adjacent regions of the MRSA strains in the present study. SCCmec type IV strains (inner four circles) showed the absence of phenol-soluble modulin-mec (psm-mec), mecI, and part of mecR in the SCCmec region, and some genes, such as cstB and cstR, for sulfide detoxification were absent when compared with the corresponding genes in the SCCmec type II strain HL21008. MRSA: methicillin-resistant Staphylococcus aureus; SCCmec: staphylococcal cassette chromosome mec.
The present study was approved by the Institutional Review Board of Kangdong Sacred Heart Hospital (NON2023-001).
No potential conflicts of interest relevant to this article were reported.
The present study was supported by the Bio and Medical Technology Development Program of the National Research Foundation (NRF), funded by the Korean government and the Ministry of Science and ICT (MSIT) 2017M3A9E4077232. The present study was supported by the Kangdong Sacred Heart Hospital Research Fund. The funders played no role in the study design, data collection, interpretation, or publication decisions.
Dataset 1. Complete genome sequence data for MRSA strains have been deposited in the NCBI database with BioProject PRJNA693997
Additional datasets generated during the current study are available from the corresponding author upon request.
1. Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler Jr. VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 2015;28:603-61.


2. Lee H, Yoon EJ, Kim D, Jeong SH, Won EJ, Shin JH, et al. Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: first one-year report from KorGLASS. Euro Surveill 2018;23:1800047.


3. Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. Atlanta, Georgia; U.S. Department of Health and Human Services, CDC: 2019.
4. Chambers HF and Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev -Microbiol 2009;7:629-41.


5. David MZ and Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 2010;23:616-87.


6. Quainoo S, Coolen JPM, van Hijum S, Huynen MA, Melchers WJG, van Schaik W, et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev 2017;30:1015-63.


7. Zhou W, Jin Y, Liu X, Chen Y, Shen P, Xiao Y. Comparison of genetic features and evolution of global and Chinese strains of community-associated methicillin-resistant Staphylococcus aureus ST22. Microbiol Spectr 2022;10:e0203721.


8. Zhou W, Jin Y, Zhou Y, Wang Y, Xiong L, Luo Q, et al. Comparative genomic analysis provides insights into the evolution and genetic diversity of community-genotype sequence type 72 Staphylococcus aureus isolates. mSystems 2021;6:e0098621.


9. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017;13:e1005595.


10. Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 1997;388:53947.

11. Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 2001;8:11-22.

12. Blattner FR, Plunkett 3rd G, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science 1997;277:1453-62.

13. Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, Enright MC, et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci U S A 2001;98:182-7.


14. Batool N, Shamim A, Chaurasia AK, Kim KK. Genome-wide analysis of Staphylococcus aureus sequence type 72 isolates provides insights into resistance against antimicrobial agents and virulence potential. Front Microbiol 2020;11:613800.


15. Takahashi T, Kim H, Kim HS, Kim HS, Song W, Kim JS. Comparative genomic analysis of staphylococcal cassette chromosome mec type V Staphylococcus aureus strains and estimation of the emergence of SCCmec V clinical isolates in Korea. Ann Lab Med 2024;44:47-55.


16. Chen Y, Chatterjee SS, Porcella SF, Yu YS, Otto M. Complete genome sequence of a Panton-Valentine leukocidin-negative community-associated methicillin-resistant Staphylococcus aureus strain of sequence type 72 from Korea. PLoS One 2013;8:e72803.


17. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 2007;51:264-74.


18. Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 2003;41:5442-8.


19. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068-9.

20. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312-3.


21. Grant JR, Arantes AS, Stothard P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics 2012;13:202.


22. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid largescale prokaryote pan genome analysis. Bioinformatics 2015;31:3691-3.


23. Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 2008;190:300-10.


24. Lee DS, Burd H, Liu J, Almaas E, Wiest O, Barabasi AL, et al. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel antimicrobial drug targets. J Bacteriol 2009;191:4015-24.


25. Schelli K, Zhong F, Zhu J. Comparative metabolomics revealing Staphylococcus aureus metabolic response to different antibiotics. Microb Biotechnol 2017;10:1764-74.


26. Vestergaard M, Leng B, Haaber J, Bojer MS, Vegge CS, Ingmer H. Genome-wide identification of antimicrobial intrinsic resistance determinants in Staphylococcus aureus. Front Microbiol 2016;7:2018.


27. Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 2013;4:e00537-12.


28. Firth N, Jensen SO, Kwong SM, Skurray RA, Ramsay JP. Staphylococcal plasmids, transposable and integrative elements. Microbiol Spectr 2018;6:10.1128.


29. Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, et al. How clonal is Staphylococcus aureus? J Bacteriol 2003;185:3307-16.


30. Qin L, McCausland JW, Cheung GY, Otto M. PSM-Mec-A virulence determinant that connects transcriptional regulation, virulence, and antibiotic resistance in staphylococci. Front Microbiol 2016;7:1293.


31. Shen J, Keithly ME, Armstrong RN, Higgins KA, Edmonds KA, Giedroc DP. Staphylococcus aureus CstB is a novel multidomain persulfide dioxygenase-sulfurtransferase involved in hydrogen sulfide detoxification. Biochemistry 2015;54:4542-54.


32. Weikum J, Ritzmann N, Jelden N, Klockner A, Herkersdorf S, Josten M, et al. Sulfide protects Staphylococcus aureus from aminoglycoside antibiotics but cannot be regarded as a general defense mechanism against antibiotics. Antimicrob Agents Chemother 2018;62:e00602-18.


33. Tam K and Torres VJ. Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol Spectr 2019;7:10.1128/microbiolspec.gpp3-0039-2018.


34. Lee SS, Kim YJ, Chung DR, Jung KS, Kim JS. Invasive infection caused by a community-associated methicillin-resistant Staphylococcus aureus strain not carrying Panton-Valentine leukocidin in South Korea. J Clin Microbiol 2010;48:311-3.


35. Kim JS, Park JS, Song W, Kim HS, Cho HC, Lee KM, et al. Panton-Valentine leukocidin positive Staphylococcus aureus isolated from blood in Korea. Korean J Lab Med 2007;27:28691.

36. Kim JS, Kim HS, Song W, Cho HC, Lee KM, Kim EC. Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates with toxic shock syndrome toxin and staphylococcal enterotoxin C genes. Korean J Lab Med 2007;27:118-23.

37. Bronesky D, Wu Z, Marzi S, Walter P, Geissmann T, Moreau K, et al. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu Rev Microbiol 2016;70:299-316.

38. Sorensen HM, Keogh RA, Wittekind MA, Caillet AR, Wiemels RE, Laner EA, et al. Reading between the lines: utilizing RNA-Seq data for global analysis of sRNAs in Staphylococcus aureus. mSphere 2020;5:e00439-20.


1. Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler Jr. VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 2015;28:603-61.


2. Lee H, Yoon EJ, Kim D, Jeong SH, Won EJ, Shin JH, et al. Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: first one-year report from KorGLASS. Euro Surveill 2018;23:1800047.


3. Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. Atlanta, Georgia; U.S. Department of Health and Human Services, CDC: 2019.
4. Chambers HF and Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev -Microbiol 2009;7:629-41.


5. David MZ and Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 2010;23:616-87.


6. Quainoo S, Coolen JPM, van Hijum S, Huynen MA, Melchers WJG, van Schaik W, et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev 2017;30:1015-63.


7. Zhou W, Jin Y, Liu X, Chen Y, Shen P, Xiao Y. Comparison of genetic features and evolution of global and Chinese strains of community-associated methicillin-resistant Staphylococcus aureus ST22. Microbiol Spectr 2022;10:e0203721.


8. Zhou W, Jin Y, Zhou Y, Wang Y, Xiong L, Luo Q, et al. Comparative genomic analysis provides insights into the evolution and genetic diversity of community-genotype sequence type 72 Staphylococcus aureus isolates. mSystems 2021;6:e0098621.


9. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017;13:e1005595.


10. Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 1997;388:53947.

11. Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 2001;8:11-22.

12. Blattner FR, Plunkett 3rd G, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science 1997;277:1453-62.

13. Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, Enright MC, et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci U S A 2001;98:182-7.


14. Batool N, Shamim A, Chaurasia AK, Kim KK. Genome-wide analysis of Staphylococcus aureus sequence type 72 isolates provides insights into resistance against antimicrobial agents and virulence potential. Front Microbiol 2020;11:613800.


15. Takahashi T, Kim H, Kim HS, Kim HS, Song W, Kim JS. Comparative genomic analysis of staphylococcal cassette chromosome mec type V Staphylococcus aureus strains and estimation of the emergence of SCCmec V clinical isolates in Korea. Ann Lab Med 2024;44:47-55.


16. Chen Y, Chatterjee SS, Porcella SF, Yu YS, Otto M. Complete genome sequence of a Panton-Valentine leukocidin-negative community-associated methicillin-resistant Staphylococcus aureus strain of sequence type 72 from Korea. PLoS One 2013;8:e72803.


17. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 2007;51:264-74.


18. Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 2003;41:5442-8.


19. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068-9.

20. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312-3.


21. Grant JR, Arantes AS, Stothard P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics 2012;13:202.


22. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid largescale prokaryote pan genome analysis. Bioinformatics 2015;31:3691-3.


23. Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 2008;190:300-10.


24. Lee DS, Burd H, Liu J, Almaas E, Wiest O, Barabasi AL, et al. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel antimicrobial drug targets. J Bacteriol 2009;191:4015-24.


25. Schelli K, Zhong F, Zhu J. Comparative metabolomics revealing Staphylococcus aureus metabolic response to different antibiotics. Microb Biotechnol 2017;10:1764-74.


26. Vestergaard M, Leng B, Haaber J, Bojer MS, Vegge CS, Ingmer H. Genome-wide identification of antimicrobial intrinsic resistance determinants in Staphylococcus aureus. Front Microbiol 2016;7:2018.


27. Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 2013;4:e00537-12.


28. Firth N, Jensen SO, Kwong SM, Skurray RA, Ramsay JP. Staphylococcal plasmids, transposable and integrative elements. Microbiol Spectr 2018;6:10.1128.


29. Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, et al. How clonal is Staphylococcus aureus? J Bacteriol 2003;185:3307-16.


30. Qin L, McCausland JW, Cheung GY, Otto M. PSM-Mec-A virulence determinant that connects transcriptional regulation, virulence, and antibiotic resistance in staphylococci. Front Microbiol 2016;7:1293.


31. Shen J, Keithly ME, Armstrong RN, Higgins KA, Edmonds KA, Giedroc DP. Staphylococcus aureus CstB is a novel multidomain persulfide dioxygenase-sulfurtransferase involved in hydrogen sulfide detoxification. Biochemistry 2015;54:4542-54.


32. Weikum J, Ritzmann N, Jelden N, Klockner A, Herkersdorf S, Josten M, et al. Sulfide protects Staphylococcus aureus from aminoglycoside antibiotics but cannot be regarded as a general defense mechanism against antibiotics. Antimicrob Agents Chemother 2018;62:e00602-18.


33. Tam K and Torres VJ. Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol Spectr 2019;7:10.1128/microbiolspec.gpp3-0039-2018.


34. Lee SS, Kim YJ, Chung DR, Jung KS, Kim JS. Invasive infection caused by a community-associated methicillin-resistant Staphylococcus aureus strain not carrying Panton-Valentine leukocidin in South Korea. J Clin Microbiol 2010;48:311-3.


35. Kim JS, Park JS, Song W, Kim HS, Cho HC, Lee KM, et al. Panton-Valentine leukocidin positive Staphylococcus aureus isolated from blood in Korea. Korean J Lab Med 2007;27:28691.

36. Kim JS, Kim HS, Song W, Cho HC, Lee KM, Kim EC. Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates with toxic shock syndrome toxin and staphylococcal enterotoxin C genes. Korean J Lab Med 2007;27:118-23.

37. Bronesky D, Wu Z, Marzi S, Walter P, Geissmann T, Moreau K, et al. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu Rev Microbiol 2016;70:299-316.

38. Sorensen HM, Keogh RA, Wittekind MA, Caillet AR, Wiemels RE, Laner EA, et al. Reading between the lines: utilizing RNA-Seq data for global analysis of sRNAs in Staphylococcus aureus. mSphere 2020;5:e00439-20.

